

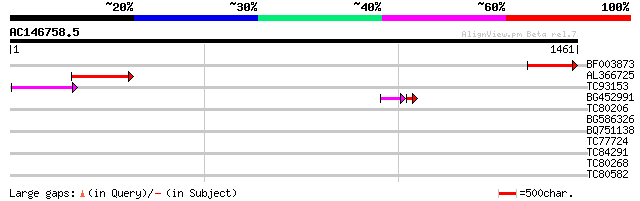
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146758.5 - phase: 0 /pseudo
(1461 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BF003873 similar to GP|14715222|em putative polyprotein {Cicer a... 183 5e-46
AL366725 154 2e-37
TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Ci... 90 6e-18
BG452991 PIR|A25875|A25 histone H4 - Tetrahymena thermophila, pa... 53 2e-07
TC80206 similar to PIR|H96764|H96764 protein RING zinc finger pr... 32 1.4
BG586326 similar to PIR|G84493|G8 probable retroelement pol poly... 32 1.8
BQ751138 weakly similar to GP|15991793|gb platelet binding prote... 31 3.1
TC77724 similar to GP|21537121|gb|AAM61462.1 photosystem II {Ara... 31 3.1
TC84291 homologue to GP|22497314|gb|AAL65624.1 transaldolase-lik... 31 3.1
TC80268 similar to GP|20152975|gb|AAM13441.1 similar to H. sapie... 30 5.3
TC80582 similar to GP|20521307|dbj|BAB91821. contains EST AU0332... 30 5.3
>BF003873 similar to GP|14715222|em putative polyprotein {Cicer arietinum},
partial (82%)
Length = 559
Score = 183 bits (464), Expect = 5e-46
Identities = 103/128 (80%), Positives = 105/128 (81%)
Frame = +3
Query: 1334 DVL*SQRS*PRSSLVRIRYWKELERWLTEWVYRRIFRICTTFSMCRNFESMFRIHLM*FR 1393
DVL*SQRS* SLVRIRY KELERW EW Y RIF IC MCRNF SMFRIHLM* R
Sbjct: 9 DVL*SQRS*L*DSLVRIRYQKELERWRIEWDYHRIF*ICMMSFMCRNFGSMFRIHLM*SR 188
Query: 1394 VTMCKLETTLR*RLYR*GLMIVK*RR*EARRYLS*ESFGRERLVKA*RGSLRVRCWSLIQ 1453
V MCK ETTLR*RLYR*GLMI K*R *EARRYL *ESFG ER+VKA*RGSLRVR WSLIQ
Sbjct: 189 VMMCKSETTLR*RLYR*GLMIEK*RH*EARRYLL*ESFGTERMVKA*RGSLRVRWWSLIQ 368
Query: 1454 NCLLEVNF 1461
+CL EVNF
Sbjct: 369 SCLHEVNF 392
>AL366725
Length = 485
Score = 154 bits (389), Expect = 2e-37
Identities = 99/160 (61%), Positives = 115/160 (71%), Gaps = 1/160 (0%)
Frame = +1
Query: 160 VLSALYC*EF*VLEMYQVRERFEARH*EGDWIPTAESFSGFG**LQDL*RGYEGSLQGSE 219
VLSAL C*+ *VLEM+QVRE FEARH EG+ IPTA SF F LQ+L GY GS QG E
Sbjct: 4 VLSALCC*DC*VLEMHQVREWFEARHQEGNRIPTAPSFP*FSEYLQNLRGGY*GS*QGCE 183
Query: 220 *KEGQGTAESS*AVQCPR*QR*TEDGRC*AA*EEGC-CRDCVFQLW*ERPQKQCLPGGNQ 278
* E QG ES *A+ CP * TE+GR *A+*EEGC C DC+FQLW ERPQ+ CL +Q
Sbjct: 184 *AEDQGPIESP*AL*CPC**GQTENGR**AS*EEGCSCGDCLFQLWRERPQE*CLS*RDQ 363
Query: 279 EVCLVWQERSCCS*LQSYRHCVL*LQWRGSH*FTVYSA*E 318
E+C V QE S CS*LQ+ +CVL LQ RGS+ F + +A*E
Sbjct: 364 EMCPV*QEGSYCS*LQAK*YCVLQLQRRGSYWFPMQAA*E 483
>TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Cicer
arietinum}, partial (8%)
Length = 516
Score = 90.1 bits (222), Expect = 6e-18
Identities = 63/170 (37%), Positives = 87/170 (51%)
Frame = +1
Query: 6 WKKRCCVSCCSTSCCPSCGTTT*RKCWCERGS*DVGDVHEEEPSDLQRTL*P*WSPDVA* 65
WK CC CC+ SC T T* W D D EE +++Q + *W +VA
Sbjct: 7 WK**CCPCCCA*GGSSSCSTAT*G*YW**WNQ-DARDFLEESSTNIQG*VCS*WCLEVAE 183
Query: 66 GD*EDLPGYAVHGGSESAIWYSSAGRGS**LVGCSSAYPWAGGSCCDLGCFQERVPEKIL 125
GD*+++P +A+ +E A+W + *LV A AG C +LG QE V ++
Sbjct: 184 GD*KNIPSHAMF*DTEGAVWDAHVS*RGR*LVD*FVACSGAG*CCGNLGHVQEGVSGQVF 363
Query: 126 SGRCSREERDRIP*AEARKYVCDRVCCQVRGAV*VLSALYC*EF*VLEMY 175
S C ERD I E +VC RVCC+V G +L +L C + *VL++Y
Sbjct: 364 SRGCQG*ERD*ISGVETG*HVCHRVCCKVCGTCHILPSLQCGDS*VLQVY 513
>BG452991 PIR|A25875|A25 histone H4 - Tetrahymena thermophila, partial (33%)
Length = 560
Score = 53.1 bits (126), Expect(2) = 2e-07
Identities = 36/65 (55%), Positives = 39/65 (59%)
Frame = +1
Query: 955 LNSLEI*VWFPNGHLRV*NWVC*RLIVNF*KVSRKHRKLM*SLWTCWLLEIRLKTVILKS 1014
LNS E *VWF RV*NW C*R NF VS++ R+ M*S L IRLK VILK
Sbjct: 70 LNSSET*VWFVKCRHRV*NWGC*RSTTNFWIVSKRLRRWM*SW*I*CLGIIRLKMVILKL 249
Query: 1015 TIKVC 1019
IK C
Sbjct: 250 MIKEC 264
Score = 21.6 bits (44), Expect(2) = 2e-07
Identities = 13/28 (46%), Positives = 20/28 (71%), Gaps = 1/28 (3%)
Frame = +3
Query: 1023 EEEFVSQTMKRL-RR*FLKRVIEVA*VF 1049
+++ V Q R+ +R*F K+VIEV *+F
Sbjct: 249 DDQGVLQFRDRI*KR*F*KKVIEVM*IF 332
>TC80206 similar to PIR|H96764|H96764 protein RING zinc finger protein
F25P22.18 [imported] - Arabidopsis thaliana, partial
(6%)
Length = 896
Score = 32.3 bits (72), Expect = 1.4
Identities = 12/31 (38%), Positives = 16/31 (50%), Gaps = 2/31 (6%)
Frame = -3
Query: 357 YWCY--ALFYCFRLCFCFGSCFV*YEWRDGC 385
YWCY + ++C CFG C V + W C
Sbjct: 330 YWCY*HSYYFCCFSLLCFGYCHVWFHWMCWC 238
>BG586326 similar to PIR|G84493|G8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 736
Score = 32.0 bits (71), Expect = 1.8
Identities = 40/115 (34%), Positives = 50/115 (42%)
Frame = +3
Query: 825 LWYIVMRPSWV*EVYLCKKVKW*LMLQDS*EFMRRIILRMISSWRL*SLY*RYGDITCMV 884
+W+I R S * Y + Q S*E MR MI W * *R+G TCMV
Sbjct: 48 MWFIQTRLSLD*VAY*PSMRRSSPTRQGS*ENMRETTPPMI*KWLR*YSP*RFGAHTCMV 227
Query: 885 RDLRCLVITRV*SICSIRRN*I*DSEDGLNC*KIMTLA*IIIQVKLMLLQMP*VG 939
R + +V*SI +* * G N * +II+ KL+ Q * G
Sbjct: 228 PRFRYIRTIKV*SIFLPSLS*T*GRGGGWNS*LTTI*TSLIIREKLIW*QTL*AG 392
>BQ751138 weakly similar to GP|15991793|gb platelet binding protein GspB
{Streptococcus gordonii}, partial (1%)
Length = 703
Score = 31.2 bits (69), Expect = 3.1
Identities = 9/18 (50%), Positives = 13/18 (72%)
Frame = +1
Query: 359 CYALFYCFRLCFCFGSCF 376
C+ +++C LCFCF CF
Sbjct: 205 CFGVYFCICLCFCFCFCF 258
>TC77724 similar to GP|21537121|gb|AAM61462.1 photosystem II {Arabidopsis
thaliana}, partial (48%)
Length = 569
Score = 31.2 bits (69), Expect = 3.1
Identities = 15/30 (50%), Positives = 16/30 (53%)
Frame = +2
Query: 3 GNGWKKRCCVSCCSTSCCPSCGTTT*RKCW 32
G WKKR V CCS+SCC C CW
Sbjct: 179 GQQWKKRLDV-CCSSSCCLLC-------CW 244
>TC84291 homologue to GP|22497314|gb|AAL65624.1 transaldolase-like protein
{Arabidopsis thaliana}, partial (25%)
Length = 821
Score = 31.2 bits (69), Expect = 3.1
Identities = 11/20 (55%), Positives = 14/20 (70%), Gaps = 1/20 (5%)
Frame = +3
Query: 354 YY*YW-CYALFYCFRLCFCF 372
YY*Y+ CY YCF C+C+
Sbjct: 609 YY*YYYCYCYCYCFSYCYCY 668
>TC80268 similar to GP|20152975|gb|AAM13441.1 similar to H. sapiens NNP-1 /
Nop52 AP001752 (Score = 92; E= 9e-18), partial (16%)
Length = 1105
Score = 30.4 bits (67), Expect = 5.3
Identities = 17/54 (31%), Positives = 28/54 (51%)
Frame = -2
Query: 1038 FLKRVIEVA*VFIRELRRCTMILRRFSGGLV*NEMWHSLCIPV*FVRSRKLSIR 1091
F+ R + F+++L I + G N++WH L IP+ +R+RKL R
Sbjct: 270 FINRTFLLLLAFLQQLTILNNISINLNNG---NQIWHFLVIPINRIRNRKLKPR 118
>TC80582 similar to GP|20521307|dbj|BAB91821. contains EST
AU033244(S5767)~unknown protein {Oryza sativa (japonica
cultivar-group)}, partial (14%)
Length = 1251
Score = 30.4 bits (67), Expect = 5.3
Identities = 14/36 (38%), Positives = 17/36 (46%), Gaps = 1/36 (2%)
Frame = -1
Query: 351 FSCYY*Y-WCYALFYCFRLCFCFGSCFV*YEWRDGC 385
F CY Y CY + C+ LC C CF + D C
Sbjct: 330 FDCYLHYILCYYILCCYILCHCILFCFHEHVLHDQC 223
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.371 0.167 0.662
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 53,161,559
Number of Sequences: 36976
Number of extensions: 911136
Number of successful extensions: 15049
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 3351
Number of HSP's successfully gapped in prelim test: 602
Number of HSP's that attempted gapping in prelim test: 10399
Number of HSP's gapped (non-prelim): 5518
length of query: 1461
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1353
effective length of database: 5,021,319
effective search space: 6793844607
effective search space used: 6793844607
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 36 (21.8 bits)
S2: 65 (29.6 bits)
Medicago: description of AC146758.5


