

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146758.11 + phase: 0 /pseudo
(1309 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
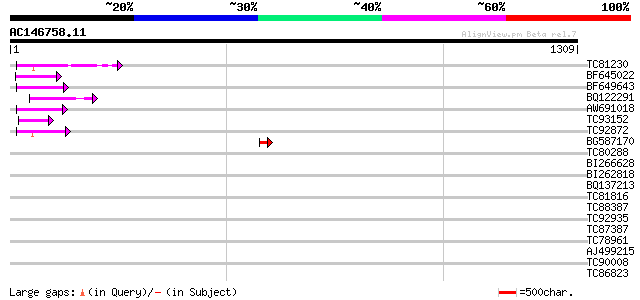
Score E
Sequences producing significant alignments: (bits) Value
TC81230 104 2e-22
BF645022 weakly similar to GP|8777581|dbj retroelement pol polyp... 65 1e-10
BF649643 similar to PIR|H86486|H86 protein Ty1/copia-element pol... 57 4e-08
BQ122291 57 6e-08
AW691018 weakly similar to GP|2462935|em open reading frame 1 {B... 55 1e-07
TC93152 49 2e-05
TC92872 43 7e-04
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 43 7e-04
TC80288 weakly similar to PIR|T06029|T06029 hypothetical protein... 43 0.001
BI266628 41 0.003
BI262818 weakly similar to GP|6642775|gb| gag-pol polyprotein {V... 41 0.003
BQ137213 40 0.008
TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana taba... 39 0.013
TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finge... 37 0.038
TC92935 weakly similar to GP|17933303|gb|AAL48234.1 At1g12650/T1... 37 0.050
TC87387 homologue to PIR|T47775|T47775 hypothetical protein F24I... 36 0.11
TC78961 similar to GP|18252179|gb|AAL61922.1 unknown protein {Ar... 36 0.11
AJ499215 weakly similar to GP|6642775|gb| gag-pol polyprotein {V... 36 0.11
TC90008 weakly similar to GP|4249376|gb|AAD14473.1| Strong simil... 35 0.15
TC86823 similar to PIR|C86390|C86390 hypothetical protein AAF985... 35 0.19
>TC81230
Length = 958
Score = 104 bits (260), Expect = 2e-22
Identities = 73/257 (28%), Positives = 119/257 (45%), Gaps = 14/257 (5%)
Frame = +1
Query: 17 SGKNYSAWEFQFRMYVKGERLWSHLNGVSKAPTEKA---------ALEEWETKDAQIITW 67
+G NY+ W ++KG RLW ++ G K PT+ LEEW++K+ QIITW
Sbjct: 241 NGSNYNHWAESMCGFLKGRRLWRYVTGDKKCPTKGKDDTADAFADKLEEWDSKNHQIITW 420
Query: 68 ILSTINPQMINNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYK-QGNSSVQEYY 126
+T P + F A+E+W++LK+ Y + + ++QL +++N K Q V E+
Sbjct: 421 FRNTSIPSIHMQFGRFENAKEVWDHLKQRYTISDLSHQYQLLKDLSNLKQQSGQPVYEFL 600
Query: 127 SGFLNLWTEHSAIIHADVPNIYLAAVQEVYNTSKQD----QFLMILRPEFEVVRGSLLNR 182
+ +W + + + P++ A + Y T + QFLM L E+E VR S L++
Sbjct: 601 AQMEVIWNQ----LTSCEPSLKDATDMKTYETHRNRVRLIQFLMALTDEYEPVRASSLHQ 768
Query: 183 NVVPSLDTCVSELLREEQRLLTQGAMSRDALIFESTTVAYAAQRRGKGHDMQQVQCFSCK 242
N +P+L+ + L EE RL L+ +A+A C C+
Sbjct: 769 NPLPTLENALPCLKSEETRL---------QLVPPKADLAFAVTNNA------TKPCRHCQ 903
Query: 243 QFGHIARSCSKKFCNYC 259
+ GH C C C
Sbjct: 904 KSGHSFSDCPTIECRNC 954
>BF645022 weakly similar to GP|8777581|dbj retroelement pol polyprotein-like
{Arabidopsis thaliana}, partial (8%)
Length = 685
Score = 65.5 bits (158), Expect = 1e-10
Identities = 30/105 (28%), Positives = 53/105 (49%)
Frame = -2
Query: 14 VRFSGKNYSAWEFQFRMYVKGERLWSHLNGVSKAPTEKAALEEWETKDAQIITWILSTIN 73
++ G NY W R + +R + + G PT LE+W+ + +I W+L+TI
Sbjct: 330 IQLRGLNYDEWSRAIRTSFQAKRKYGFVEGKIPKPTTPEKLEDWKAVQSMLIAWLLNTIE 151
Query: 74 PQMINNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQG 118
P + + L + A+ +W +LK+ + N A+ QL+ + KQG
Sbjct: 150 PSLRSTLSYYDDAESLWTHLKQRFCVVNGARICQLKASLGECKQG 16
>BF649643 similar to PIR|H86486|H86 protein Ty1/copia-element polyprotein
[imported] - Arabidopsis thaliana, partial (2%)
Length = 608
Score = 57.4 bits (137), Expect = 4e-08
Identities = 29/124 (23%), Positives = 56/124 (44%), Gaps = 3/124 (2%)
Frame = +1
Query: 15 RFSGKNYSAWEFQFRMYVKGERLWSHLNGVSKAPTE---KAALEEWETKDAQIITWILST 71
+ +G NYS+W + + +NG K P+E A W ++ I++W+ +
Sbjct: 172 KLNGTNYSSWSRSMVHALTAKNKVGFINGSIKTPSEVDQPAEYALWNRCNSMILSWLTHS 351
Query: 72 INPQMINNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQEYYSGFLN 131
+ P + + A ++W K ++Q N +Q++ +A+ QG S Y++
Sbjct: 352 VEPDLAKGVIHAKTACQVWEDFKDQFSQKNIPAIYQIQKSLASLSQGTVSASTYFTKIKG 531
Query: 132 LWTE 135
LW E
Sbjct: 532 LWDE 543
>BQ122291
Length = 487
Score = 56.6 bits (135), Expect = 6e-08
Identities = 40/160 (25%), Positives = 73/160 (45%), Gaps = 4/160 (2%)
Frame = -2
Query: 46 KAPTEKAALEEWETKDAQIITWILSTINPQMINNLRSFSFAQEMWNYLKRIYNQDNPAKR 105
+A T A EWE KD+ + TWILS I+P +++ + ++W+ + +
Sbjct: 474 EAGTRNPAYTEWEDKDSLLCTWILSRISPSLLSRFVLLRHSWQVWDEIHSYCFTQMKTRS 295
Query: 106 FQLELEIANYKQGNSSVQEYYSGFL----NLWTEHSAIIHADVPNIYLAAVQEVYNTSKQ 161
QL E+ + +G+ +V E+ + +L + + H D+ + L A+ E
Sbjct: 294 RQLRSELRSITKGSRTVSEFIARIRAISESLASIGDPVSHRDLIEVVLEALPE------- 136
Query: 162 DQFLMILRPEFEVVRGSLLNRNVVPSLDTCVSELLREEQR 201
EF+ + S+ ++ V SLD S+LL +E R
Sbjct: 135 ---------EFDPIVASVNAKSEVVSLDELESQLLTQESR 43
>AW691018 weakly similar to GP|2462935|em open reading frame 1 {Brassica
oleracea}, partial (12%)
Length = 639
Score = 55.5 bits (132), Expect = 1e-07
Identities = 28/117 (23%), Positives = 60/117 (50%), Gaps = 1/117 (0%)
Frame = +2
Query: 17 SGKNYSAWEFQFRMYVKGERLWSHLNGVSKAPT-EKAALEEWETKDAQIITWILSTINPQ 75
+G NY W + + + ++G KAP+ + L W+ + ++TWIL +I P
Sbjct: 212 NGGNYGEWSRSMLLSLSAKNKLGLIDGTVKAPSADDPKLPLWKRCNDLVLTWILHSIEPD 391
Query: 76 MINNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQEYYSGFLNL 132
+ ++ A +W+ L ++Q + ++ +Q+ EI+ +QG+ + +YY+ +L
Sbjct: 392 IARSVIFSDTAAAVWSDLHDRFSQGDESRIYQIRQEISECRQGSLLISDYYTKLKSL 562
>TC93152
Length = 647
Score = 48.5 bits (114), Expect = 2e-05
Identities = 23/83 (27%), Positives = 39/83 (46%), Gaps = 2/83 (2%)
Frame = +3
Query: 20 NYSAWEFQFRMYVKGERLWSHLNGVSKA--PTEKAALEEWETKDAQIITWILSTINPQMI 77
NY W R Y+ G+ LW + +S++ P + E W ++ + + I P+ I
Sbjct: 162 NYDYWSCLVRNYLLGQDLWGFVTSISESTGPRSRRETEVWNRRNGKALHIIQLACGPENI 341
Query: 78 NNLRSFSFAQEMWNYLKRIYNQD 100
N ++ A+E WN L Y+ D
Sbjct: 342 NLIKDLQSAREAWNELSTHYSSD 410
>TC92872
Length = 923
Score = 43.1 bits (100), Expect = 7e-04
Identities = 29/135 (21%), Positives = 58/135 (42%), Gaps = 10/135 (7%)
Frame = +1
Query: 15 RFSGKNYSAWEFQFRMYVKGERLWSHLNGVSKAPTE----------KAALEEWETKDAQI 64
+ S KN+ AW+ Q + G + H++G + TE +W T D I
Sbjct: 106 KLSRKNFRAWKRQVTTLLAGIEVMGHIDGTTPIQTETIINNGVSAPNPDYTKWFTLDQLI 285
Query: 65 ITWILSTINPQMINNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSSVQE 124
I +LS++ + S+ A+ +W ++ +N + + + +I +G+ S+ +
Sbjct: 286 INLLLSSMTEADSISFASYETARTLWVAIESQFNNTSRSHVMSVTNQIQRCTKGDKSITD 465
Query: 125 YYSGFLNLWTEHSAI 139
Y +L E + I
Sbjct: 466 YLFSVKSLADELAVI 510
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 43.1 bits (100), Expect = 7e-04
Identities = 18/31 (58%), Positives = 23/31 (74%)
Frame = -3
Query: 577 TDSGGEYMSHEFQEYLQHKGILSQ*SCPNTP 607
+D+GGEY S+ F+ +L H GIL Q SCP TP
Sbjct: 230 SDNGGEYTSYAFKSHLDHHGILHQTSCPYTP 138
>TC80288 weakly similar to PIR|T06029|T06029 hypothetical protein T28I19.100
- Arabidopsis thaliana, partial (14%)
Length = 1460
Score = 42.7 bits (99), Expect = 0.001
Identities = 42/133 (31%), Positives = 64/133 (47%), Gaps = 20/133 (15%)
Frame = -1
Query: 652 LIIFLLWFLTLTLLSFVF---SKLILIIVICILLGVCVLFIYLRLKGISLE----HNLFS 704
L++F+L L +TL FVF LIL I+IC+ + ++FIYL + +F
Sbjct: 758 LLVFILR-LIITLFFFVFLFIHNLILTIIICL*F-LMIIFIYLNISSFIFHLFTFAFIFL 585
Query: 705 VHLWGIVTFIRDL----------CFMMSLIIASEFRGMLHFLIINSCFILFLLT*M-ILL 753
+H ++ F L F S ++ S F +L F++ FIL ++ M I L
Sbjct: 584 LHFCSMLIFFFILFFSPFMHIVFTFFFSFVLVSFFSFLLLFIVFT--FILHIVYNMFIFL 411
Query: 754 FF--PIFLLCLNL 764
FF P L+C NL
Sbjct: 410 FFLMPFILICFNL 372
>BI266628
Length = 536
Score = 41.2 bits (95), Expect = 0.003
Identities = 23/72 (31%), Positives = 39/72 (53%)
Frame = -1
Query: 46 KAPTEKAALEEWETKDAQIITWILSTINPQMINNLRSFSFAQEMWNYLKRIYNQDNPAKR 105
+A T+K A EWE +D+ + TWILSTI P +++ F + W ++ I AK
Sbjct: 299 EAGTKKPAYTEWEEQDSLLCTWILSTI*PSLLSR---FVLLRHSWFGMRFIVTASCKAKI 129
Query: 106 FQLELEIANYKQ 117
L + A++++
Sbjct: 128 LALPIVSASHER 93
>BI262818 weakly similar to GP|6642775|gb| gag-pol polyprotein {Vitis
vinifera}, partial (24%)
Length = 615
Score = 41.2 bits (95), Expect = 0.003
Identities = 27/130 (20%), Positives = 59/130 (44%), Gaps = 13/130 (10%)
Frame = +1
Query: 16 FSGKNYSAWEFQFRMYVKGERLWSHLNG------VSKAPTEKAAL--EEWETKDAQIITW 67
F G+NY W + Y++ LW + +S PT +E +T+ ++
Sbjct: 184 FDGENYHIWAARIEAYLEANDLWEAVEEDYEVLPLSDNPTMAQIKNHKERKTRKSKARAT 363
Query: 68 ILSTINPQMINNLRSFSFAQEMWNYLKRIYNQDNPAKRFQL-----ELEIANYKQGNSSV 122
+ + ++ ++ + + A E+WN+LK Y D + Q E E+ K+ + ++
Sbjct: 364 LFAAVSEEIFTRIMTIKSAFEIWNFLKTEYEGDERIRGMQALNLIREFEMQKMKE-SETI 540
Query: 123 QEYYSGFLNL 132
+EY + +++
Sbjct: 541 KEYANKLISI 570
>BQ137213
Length = 1006
Score = 39.7 bits (91), Expect = 0.008
Identities = 32/108 (29%), Positives = 51/108 (46%)
Frame = +2
Query: 656 LLWFLTLTLLSFVFSKLILIIVICILLGVCVLFIYLRLKGISLEHNLFSVHLWGIVTFIR 715
LL++L + LL +FS L I+IC VC+L + L + HL+ ++ R
Sbjct: 677 LLYYLLIYLLCVIFSFTSLFILICFFYSVCILLSCSMYSALLL*Y----YHLYTVLHVCR 844
Query: 716 DLCFMMSLIIASEFRGMLHFLIINSCFILFLLT*MILLFFPIFLLCLN 763
F +S+ + F HFLI F LL+ + L+F + L L+
Sbjct: 845 S-TFYLSISVFCFFLLRFHFLISALFFYAALLS--VSLYFVLCLTALS 979
>TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana tabacum},
partial (13%)
Length = 663
Score = 38.9 bits (89), Expect = 0.013
Identities = 33/108 (30%), Positives = 54/108 (49%)
Frame = -2
Query: 655 FLLWFLTLTLLSFVFSKLILIIVICILLGVCVLFIYLRLKGISLEHNLFSVHLWGIVTFI 714
F L+FL L LL F+F LIL+ + IL + V + L L + L LF++ ++ + +
Sbjct: 530 FTLFFLILLLL-FLFLILILLFLTSILTFIIVFILVLFLLLVFLRFLLFTILVFVFFSLL 354
Query: 715 RDLCFMMSLIIASEFRGMLHFLIINSCFILFLLT*MILLFFPIFLLCL 762
F++ LI+ + F +N F+LF LL F + +L L
Sbjct: 353 LLFLFLLLLILLFI---TIFFFTLNILFLLF----FFLLLFVLLILTL 231
Score = 36.2 bits (82), Expect = 0.085
Identities = 39/129 (30%), Positives = 63/129 (48%)
Frame = -2
Query: 429 FLFSLFPISYLLLVTMF*ILMRTGIENWVIQTLLFCLIYLNLVYWGLKMLFLMPLCHVLY 488
FL L +L+L+ +F T I ++I +L +++L LV+ L+ L L V +
Sbjct: 518 FLILLLLFLFLILILLF----LTSILTFIIVFIL--VLFLLLVF--LRFLLFTILVFVFF 363
Query: 489 AN*PKLKHFLSRLVLIVLLVVLR*FIMMCGVCHL*FLMLAINILSHLLMIKFVLLGYIFF 548
+ L FL L+LI+L + + F + FL+L + ++ L+ I FVLL I
Sbjct: 362 S---LLLLFLFLLLLILLFITIFFFTLNILFLLFFFLLLFVLLILTLIYISFVLLLLILL 192
Query: 549 DPSLKCFLY 557
L FLY
Sbjct: 191 FLFLIIFLY 165
>TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finger protein
{Oryza sativa (japonica cultivar-group)}, partial (96%)
Length = 1286
Score = 37.4 bits (85), Expect = 0.038
Identities = 17/40 (42%), Positives = 26/40 (64%), Gaps = 3/40 (7%)
Frame = +1
Query: 238 CFSCKQFGHIARSC-SKKFCNYCKQRGHIITECYV--RPP 274
C++CK+ GH+A SC ++ C+ C + GH EC V +PP
Sbjct: 538 CWNCKEPGHMASSCPNEGICHTCGKAGHRARECTVPQKPP 657
Score = 37.4 bits (85), Expect = 0.038
Identities = 17/39 (43%), Positives = 21/39 (53%), Gaps = 3/39 (7%)
Frame = +1
Query: 234 QQVQCFSCKQFGHIARSC---SKKFCNYCKQRGHIITEC 269
+ V C SC+QFGH++R C C C RGH EC
Sbjct: 901 RDVVCRSCQQFGHMSRDCMGGPLMICQNCGGRGHQAYEC 1017
Score = 36.2 bits (82), Expect = 0.085
Identities = 14/33 (42%), Positives = 22/33 (66%), Gaps = 1/33 (3%)
Frame = +1
Query: 238 CFSCKQFGHIARSCS-KKFCNYCKQRGHIITEC 269
C +C + GHIA C+ +K CN C++ GH+ +C
Sbjct: 673 CNNCYKQGHIAVECTNEKACNNCRKTGHLARDC 771
Score = 35.4 bits (80), Expect = 0.15
Identities = 15/33 (45%), Positives = 19/33 (57%), Gaps = 1/33 (3%)
Frame = +1
Query: 238 CFSCKQFGHIARSCS-KKFCNYCKQRGHIITEC 269
C +C GHIA CS K C CK+ GH+ + C
Sbjct: 481 CHNCSLPGHIASECSTKSLCWNCKEPGHMASSC 579
Score = 35.0 bits (79), Expect = 0.19
Identities = 15/40 (37%), Positives = 21/40 (52%), Gaps = 8/40 (20%)
Frame = +1
Query: 238 CFSCKQFGHIARSCSK--------KFCNYCKQRGHIITEC 269
C +C + GH AR C+ + CN C ++GHI EC
Sbjct: 595 CHTCGKAGHRARECTVPQKPPGDLRLCNNCYKQGHIAVEC 714
Score = 33.9 bits (76), Expect = 0.42
Identities = 18/45 (40%), Positives = 23/45 (51%), Gaps = 1/45 (2%)
Frame = +1
Query: 226 RRGKGHDMQQVQCFSCKQFGHIARSC-SKKFCNYCKQRGHIITEC 269
RRG D C +CK+ GH R C + C+ C GHI +EC
Sbjct: 397 RRGFSQDNL---CKNCKRPGHYVRECPNVAVCHNCSLPGHIASEC 522
>TC92935 weakly similar to GP|17933303|gb|AAL48234.1 At1g12650/T12C24_1
{Arabidopsis thaliana}, partial (45%)
Length = 725
Score = 37.0 bits (84), Expect = 0.050
Identities = 47/144 (32%), Positives = 64/144 (43%), Gaps = 9/144 (6%)
Frame = -2
Query: 977 IHLYLFIAHL*ELFYFFCTLMIWLLLVLIMLPFRDL*NSYKPLFI*KI--LVSCIIFLVL 1034
I LFI LFYF TL I+ + LPF+ L S K +FI*KI L II
Sbjct: 595 IRFQLFINPCYALFYFTSTLWIF---IFSQLPFKSLSFSRKIIFI*KIISLSKPIIVKCT 425
Query: 1035 RFTLHPRVSFSISTSMS*I*FPWLVSNRLI----QW-ILLLRLMLNIIVMMVVSCL--IR 1087
L +++++ S++ I NRL+ W IL+ L I + C+ IR
Sbjct: 424 AKRLKSWITYTLLRSLNDI---SKSRNRLLATSFHWPILISPFQLLIFCRFLEKCMRPIR 254
Query: 1088 YCIDNLWVVLII*LLLVLTYLLLF 1111
C I *L L L + L+F
Sbjct: 253 SCFLQFLKCYIS*LFLYLFFFLIF 182
>TC87387 homologue to PIR|T47775|T47775 hypothetical protein F24I3.230 -
Arabidopsis thaliana, partial (14%)
Length = 1074
Score = 35.8 bits (81), Expect = 0.11
Identities = 33/107 (30%), Positives = 52/107 (47%), Gaps = 5/107 (4%)
Frame = -2
Query: 654 IFLLWFLTLTLLSFVFSKLILIIVI-CILLGVCVLFIYLRLKGISLEHNLFSVHLWGIVT 712
IF+ FL L L S + +I + I I+ G V ++L L+ + F+ L +
Sbjct: 707 IFVFLFLVLLLTSISIALIISVTDIGSIIFGFFVFLLFLLLRLVHFRRTNFTRFLVLLFL 528
Query: 713 FIRDLCF---MMSLIIASEFRGMLHFLIIN-SCFILFLLT*MILLFF 755
IR L F + +L + F + + L+I S ILF+L +LLFF
Sbjct: 527 LIRLLHFRRTIFTLFLVLLFLLLFNLLLIRCSSSILFILLLFLLLFF 387
>TC78961 similar to GP|18252179|gb|AAL61922.1 unknown protein {Arabidopsis
thaliana}, partial (71%)
Length = 974
Score = 35.8 bits (81), Expect = 0.11
Identities = 19/50 (38%), Positives = 23/50 (46%), Gaps = 4/50 (8%)
Frame = +2
Query: 228 GKGHDMQQVQCFSCKQFGHIARSCS----KKFCNYCKQRGHIITECYVRP 273
G+G +CF+C GH AR C K C C +RGHI C P
Sbjct: 329 GRGPPPGSGRCFNCGIDGHWARDCKAGDWKNKCYRCGERGHIEKNCKNSP 478
>AJ499215 weakly similar to GP|6642775|gb| gag-pol polyprotein {Vitis
vinifera}, partial (18%)
Length = 567
Score = 35.8 bits (81), Expect = 0.11
Identities = 22/77 (28%), Positives = 42/77 (53%), Gaps = 7/77 (9%)
Frame = +3
Query: 327 WFLDSGASYHMTGSLEYLHNLHSYDGNKKIQIADGNTLSITDVGDI----NSDFR---DV 379
W +DSG ++HMT + L+ K+++ +G + + +G + +S ++ +V
Sbjct: 108 WLIDSGCTHHMTHDRDLFKELNK-STISKVRMLNGAHIEVEGIGTVLVKSHSGYKQISNV 284
Query: 380 LVSPGLASNLLSVGQLV 396
L +P L +LLSV QL+
Sbjct: 285 LYAPKLNQSLLSVPQLL 335
>TC90008 weakly similar to GP|4249376|gb|AAD14473.1| Strong similarity to
GP|2829864 F3I6.6 zinc metalloproteinase homolog from
Arabidopsis thaliana BAC, partial (21%)
Length = 1173
Score = 35.4 bits (80), Expect = 0.15
Identities = 33/134 (24%), Positives = 57/134 (41%), Gaps = 12/134 (8%)
Frame = +2
Query: 62 AQIITWILSTINPQMINNLRSFSFAQEMWNYLKRIYNQDNPAKRFQLELEIANYKQGNSS 121
A I I+ + P + + S +WN LK I+N L+I + G S
Sbjct: 128 APAIGLIVRSRIPFYLKRITSSKLFSSIWNKLKSIFNN---------ALKIGDIASGISD 280
Query: 122 VQEY--YSGFLNLWTEHSAIIHADVPNIYLAAVQEVYNTS-------KQDQFLMILRPEF 172
++EY + G+LN T HS A N+ A ++ N + QD + +I +P
Sbjct: 281 LKEYLQFFGYLNSSTLHSNFTDAFTENLQSAIIEFQTNFNLNTTGQLDQDIYKIISKPRC 460
Query: 173 ---EVVRGSLLNRN 183
+++ G+ +N
Sbjct: 461 GVPDIINGTTTMKN 502
>TC86823 similar to PIR|C86390|C86390 hypothetical protein AAF98579.1
[imported] - Arabidopsis thaliana, partial (13%)
Length = 659
Score = 35.0 bits (79), Expect = 0.19
Identities = 20/64 (31%), Positives = 36/64 (56%)
Frame = +3
Query: 1046 ISTSMS*I*FPWLVSNRLIQWILLLRLMLNIIVMMVVSCLIRYCIDNLWVVLII*LLLVL 1105
IS + +* W++ RL W+L + N+I M ++ + C+ N +++ I L+LVL
Sbjct: 438 ISVKIQVV*TKWVMYYRL--WLLGYKFFKNVIF*MCLTLITELCLSNKFLLSIAHLVLVL 611
Query: 1106 TYLL 1109
Y+L
Sbjct: 612 YYML 623
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.347 0.154 0.508
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 48,416,796
Number of Sequences: 36976
Number of extensions: 849852
Number of successful extensions: 11060
Number of sequences better than 10.0: 75
Number of HSP's better than 10.0 without gapping: 4758
Number of HSP's successfully gapped in prelim test: 776
Number of HSP's that attempted gapping in prelim test: 5685
Number of HSP's gapped (non-prelim): 6530
length of query: 1309
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1201
effective length of database: 5,021,319
effective search space: 6030604119
effective search space used: 6030604119
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC146758.11


