

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146757.4 - phase: 0 /pseudo
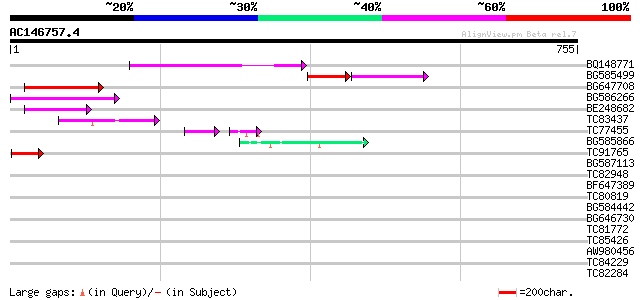
(755 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BQ148771 134 1e-31
BG585499 74 5e-29
BG647708 weakly similar to GP|13786450|gb| putative reverse tran... 89 8e-18
BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-li... 83 3e-16
BE248682 similar to GP|18568269|gb putative gag-pol polyprotein ... 58 2e-08
TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imp... 56 6e-08
TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarot... 44 7e-08
BG585866 50 2e-06
TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative ret... 47 2e-05
BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At... 41 0.001
TC82948 41 0.002
BF647389 40 0.003
TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non... 31 0.003
BG584442 39 0.007
BG646730 34 0.18
TC81772 30 0.42
TC85426 similar to SP|O23758|NLTP_CICAR Nonspecific lipid-transf... 31 1.5
AW980456 29 5.9
TC84229 weakly similar to GP|9294175|dbj|BAB02077.1 cytochrome p... 29 5.9
TC82284 weakly similar to GP|11119510|gb|AAF37724.2 diaphanous p... 26 6.6
>BQ148771
Length = 680
Score = 134 bits (338), Expect = 1e-31
Identities = 74/236 (31%), Positives = 114/236 (47%)
Frame = -3
Query: 160 KIQSKLNGWKQQCLTLAGRITLAKSVISTIPYYHMQYAKIPKTICDEIDKIQRGFIWGDS 219
++ L WK L+LA R+TLAKSVI +P Y M IPK +EI K+QR F+WGD+
Sbjct: 588 QVHVMLANWKANHLSLARRVTLAKSVIEAVPLYPMMTTIIPKACIEEIQKLQRKFVWGDT 409
Query: 220 DQGRKAHLISWDVCCLPKADGGLGFRQTHKMNEAFLMKILWNLIKNPEDLWCRVLRSKYG 279
+ R+ H + W+ PK GLG R+ MN+A +MK+ W++ L V+R KY
Sbjct: 408 EVSRRYHAVGWETMSKPKTIYGLGLRRLDVMNKACIMKLGWSIYSGSNSLCTEVMRGKYQ 229
Query: 280 RNNDLIASINAHPYDSPLWKALVNIWNDFKGHVVWNIGDGRNTNFWLDKWVPNNESLMSI 339
R+ L P DS LWKALV +W + +
Sbjct: 228 RSESLEEIFLEKPTDSSLWKALVKLWPEIE------------------------------ 139
Query: 340 GNQTYMDITLSVRDVLSPSGDWNIDFLINNLPTNTVNQILALSIPNDDDGPDTIGW 395
R+++ +G+WN + L LP + + +I+A++ P+++ G D + W
Sbjct: 138 ------------RNLVDSNGNWNWEKLKQWLPFDVLQRIMAIATPHENLGSDKLIW 7
>BG585499
Length = 792
Score = 73.9 bits (180), Expect(2) = 5e-29
Identities = 31/58 (53%), Positives = 42/58 (71%)
Frame = +3
Query: 397 GTNTHHFTVQSAYSLQHQDCPILEGDWKSIWKWHGPHRIQTFIWLDTHERILTNFQRS 454
GT+TH F ++S+Y+L D I++ DWK +W W GPHR QTF+WL H ILTN++RS
Sbjct: 147 GTSTHQFKIKSSYNLLV*DQSIVDCDWKMLWGWRGPHRTQTFMWLVAHGCILTNYRRS 320
Score = 72.8 bits (177), Expect(2) = 5e-29
Identities = 50/103 (48%), Positives = 58/103 (55%), Gaps = 1/103 (0%)
Frame = +1
Query: 456 GVVEYLLIARIVEMKMKLFFMCFAIASTLPRYGFTSFHMIL*LISF-LSIAGIGSSITSS 514
GV + L +VEM KLFFMCF IA RYG +I *LISF L IAGIG S T +
Sbjct: 325 GVRGFWLHVPVVEMLTKLFFMCFVIAGQRLRYGLDLCLLIG*LISFPLMIAGIGFSKTLA 504
Query: 515 RKTLGRTL*VGRQLS*QHVGTCGSGGIKLFLKLILLGRTTQFL 557
+ + G QLS* HVG CG G KLFLK + TQ +
Sbjct: 505 KDRMEFLSLSGSQLS*LHVGICGLGETKLFLKKDFKDQITQLM 633
>BG647708 weakly similar to GP|13786450|gb| putative reverse transcriptase
{Oryza sativa}, partial (9%)
Length = 708
Score = 88.6 bits (218), Expect = 8e-18
Identities = 45/105 (42%), Positives = 69/105 (64%)
Frame = +1
Query: 20 RGIRQGDPLSPYLFVICMDRLSHMIADQVEAKYWIPMRAGRYGPQISHLLFADDLLLFAE 79
+G+RQGDPLSPYLF++C + LS ++ + + ++ R P+I+HLLFADD LLFA
Sbjct: 10 KGLRQGDPLSPYLFILCANVLSGLLKREGNKQNLHGIQVARSDPKITHLLFADDSLLFAR 189
Query: 80 ASIEQAHCVLHCLDIFCQASGQKINRNKTCVYFSKNVDTQLREDI 124
A++ +A ++ L + ASGQ +N K+ V +S+NV Q +E I
Sbjct: 190 ANLTEAATIMQVLHSYQSASGQLVNFEKSEVSYSQNVPNQEKEMI 324
>BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-like protein
{Arabidopsis thaliana}, partial (18%)
Length = 789
Score = 83.2 bits (204), Expect = 3e-16
Identities = 46/146 (31%), Positives = 73/146 (49%), Gaps = 1/146 (0%)
Frame = -3
Query: 2 IEYIIMWNGEKTDTFFSTRGIRQGDPLSPYLFVICMDRLSHMIADQVEAKYWIPMRAGRY 61
+ Y + NG +RG+RQGDPLSPYLF++C + LS + + ++ R
Sbjct: 442 VSYSFLINGGPQGRVLPSRGLRQGDPLSPYLFILCTEVLSGLCQQALRKGTLPGVKVARN 263
Query: 62 GPQISHLLFADDLLLFAEASIEQAHCVLHCLDIFCQASGQKINRNKTCVYFSKNVDTQLR 121
P I+HLLFADD + F +++ +L +D + ASG+ IN K+ + FS +
Sbjct: 262 CPPINHLLFADDTMFFGKSNASSCAILLSIMDKYRAASGRCIN*TKSAITFSSKTSQAII 83
Query: 122 EDILHHTGFNQVNSLGMYLG-ANLAP 146
+ + + G YLG N+ P
Sbjct: 82 DRVKGELKIAKEGGTGKYLGYRNILP 5
>BE248682 similar to GP|18568269|gb putative gag-pol polyprotein {Zea mays},
partial (1%)
Length = 441
Score = 57.8 bits (138), Expect = 2e-08
Identities = 29/89 (32%), Positives = 51/89 (56%)
Frame = +3
Query: 20 RGIRQGDPLSPYLFVICMDRLSHMIADQVEAKYWIPMRAGRYGPQISHLLFADDLLLFAE 79
RG++QGDPL+P+LF++ + +S ++ + V + R G ++SHL +ADD L
Sbjct: 24 RGLKQGDPLAPFLFLLVAEGISGLMKNAVNRNLFQGFDVKRGGTRVSHLQYADDTLCIGM 203
Query: 80 ASIEQAHCVLHCLDIFCQASGQKINRNKT 108
+++ + L F ASG K+N +K+
Sbjct: 204 PTVDNLWTLKALLQGFEMASGLKVNFHKS 290
>TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imported] -
Arabidopsis thaliana, partial (6%)
Length = 951
Score = 55.8 bits (133), Expect = 6e-08
Identities = 42/139 (30%), Positives = 69/139 (49%), Gaps = 4/139 (2%)
Frame = +2
Query: 65 ISHLLFADDLLLFAEASIEQAHCVLHCLDIFCQASGQKINRNKT---CVYFSKNVDTQLR 121
+SHL FA+D LL + + L IF SG K+N +K+ CV + + ++
Sbjct: 542 VSHLQFANDTLLLETKNWANIRALRAALVIF*AMSGLKVNFHKSGLVCVNIAPSWLSEAA 721
Query: 122 EDILHHTGFNQVNSLGMYLGANLAPGRSLRGKF-NHIVNKIQSKLNGWKQQCLTLAGRIT 180
+ G LGM + G S R F IVN+I+++L GW + L+ GR+
Sbjct: 722 SVLSWKVGKVPFLYLGMPI-----EGNSRRLSFWEPIVNRIKARLTGWNSRFLSFGGRLV 886
Query: 181 LAKSVISTIPYYHMQYAKI 199
L KSV++++ Y + +K+
Sbjct: 887 LLKSVLTSLSVYALPSSKL 943
>TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarotenoid
dioxygenase1 {Pisum sativum}, partial (43%)
Length = 1865
Score = 43.5 bits (101), Expect(2) = 7e-08
Identities = 19/46 (41%), Positives = 25/46 (54%)
Frame = -3
Query: 234 CLPKADGGLGFRQTHKMNEAFLMKILWNLIKNPEDLWCRVLRSKYG 279
CLP+ GGLG R +N + L K W L+++ LW VL YG
Sbjct: 969 CLPRCKGGLGVRDIRLVNVSLLAKWWWRLLQDQSSLWKEVLEDIYG 832
Score = 31.6 bits (70), Expect(2) = 7e-08
Identities = 20/53 (37%), Positives = 27/53 (50%), Gaps = 10/53 (18%)
Frame = -2
Query: 293 YDSPLWKALVNIWNDFKGHVVW-------NIGDGRNTNFWLDKW---VPNNES 335
Y S WK L+++ + G V W +GDGR++ FW D W VP ES
Sbjct: 784 YASRWWKDLMSL--EEVGRVRWFPRELIRKVGDGRSSFFWKDAWDSSVPLRES 632
>BG585866
Length = 828
Score = 50.4 bits (119), Expect = 2e-06
Identities = 46/180 (25%), Positives = 72/180 (39%), Gaps = 8/180 (4%)
Frame = +3
Query: 306 NDFKGHVVWNIGDGRNTNFWLDKWVPNNESLMSIGNQTYM----DITLSVRDVLSPSGDW 361
N K W G G N++FW W SL +G Q D+ L+V+DV + G
Sbjct: 99 NVLKSGYTWRAGSG-NSSFWYTNW----SSLGLLGTQAPFVDIHDLHLTVKDVFTTGGQ- 260
Query: 362 NIDFLINNLPTNTVNQILALSIPNDDDGPDTIGWGGTNTHHFTVQSAYSL---QHQDCPI 418
+ L LPT+ I + + D W + +T +S YS Q +
Sbjct: 261 HTQSLYTILPTDIAEVINNTHLNFNASIGDAYIWPHNSNGVYTAKSGYSWILSQTETVNY 440
Query: 419 LEGDWKSIWKWHGPHRIQTFIWLDTHERILT-NFQRSNGVVEYLLIARIVEMKMKLFFMC 477
W IW+ P + + F+WL H + T + +V + +R E + + FF C
Sbjct: 441 NNSSWSWIWRLKIPEKYKFFLWLACHNAVPTLSLLNHRNMVNSAICSRCGEHE-ESFFHC 617
>TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative retroelement
{Oryza sativa (japonica cultivar-group)}, partial (1%)
Length = 625
Score = 47.4 bits (111), Expect = 2e-05
Identities = 18/42 (42%), Positives = 30/42 (70%)
Frame = +2
Query: 3 EYIIMWNGEKTDTFFSTRGIRQGDPLSPYLFVICMDRLSHMI 44
+Y ++ N + D +RG++QGD LSPY+F+IC++ LS +I
Sbjct: 119 DYYVLVNNDAVDPIIPSRGLQQGDHLSPYIFIICVEGLSFLI 244
>BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (10%)
Length = 767
Score = 41.2 bits (95), Expect = 0.001
Identities = 38/176 (21%), Positives = 72/176 (40%), Gaps = 27/176 (15%)
Frame = -3
Query: 292 PYDSPL-------WKALVNIWNDFKGHVVWNIGDGRNTNFWLDKWV---------PNNES 335
P ++PL W+++ + + K IG+G NTN W + PN S
Sbjct: 765 PLNAPLGSWASYAWRSIHSAQHLIKQGAKVIIGNGENTNIWEREMAWKLTCVTNHPNKHS 586
Query: 336 LMSIGNQTYMDITLSVRDVLSPSGDWNIDFLINNLPTNTVNQILALSIPNDDDGPDTIGW 395
+ T R + N + + + P T +IL++ P G D+ W
Sbjct: 585 SRAY*APTLYGYE-GCRSDDPMRRERNANLINSIFPEGTRRKILSIH-PQGPIGEDSYSW 412
Query: 396 GGTNTHHFTVQSAYSLQHQ-----------DCPILEGDWKSIWKWHGPHRIQTFIW 440
+ + H++V+S Y +Q D P L+ ++ +WK++ +++ F+W
Sbjct: 411 EYSKSGHYSVKSGYYVQTNIIAAANQRGTVDQPSLDDLYQRVWKYNTSPKVRHFLW 244
>TC82948
Length = 705
Score = 40.8 bits (94), Expect = 0.002
Identities = 17/48 (35%), Positives = 28/48 (57%)
Frame = +3
Query: 228 ISWDVCCLPKADGGLGFRQTHKMNEAFLMKILWNLIKNPEDLWCRVLR 275
+SW+ C P +G LG R K+NEA +K+ W+++ + E W +R
Sbjct: 267 VSWEKVCRPIKEGSLGIRSLSKLNEALNLKLCWDMMISKEQ-WYAFMR 407
>BF647389
Length = 404
Score = 40.0 bits (92), Expect = 0.003
Identities = 28/104 (26%), Positives = 43/104 (40%)
Frame = +2
Query: 158 VNKIQSKLNGWKQQCLTLAGRITLAKSVISTIPYYHMQYAKIPKTICDEIDKIQRGFIWG 217
V KI ++ + L+ AG + L K V+ + Y Q +P + I R F+
Sbjct: 92 VKKIICRIEN*SSKLLSYAGSLQLIKIVLFGVQPYWSQVFVLP*KVIKLIQTTCRIFL*T 271
Query: 218 DSDQGRKAHLISWDVCCLPKADGGLGFRQTHKMNEAFLMKILWN 261
K LI+ + CLPK GG N+ + K+ WN
Sbjct: 272 GKSGTSKRALIAREHICLPKTAGGWNVIDLKVXNQTAICKLXWN 403
>TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non-LTR
retroelement reverse transcriptase {Oryza sativa
(japonica cultivar-group)}, partial (2%)
Length = 1262
Score = 30.8 bits (68), Expect(2) = 0.003
Identities = 13/29 (44%), Positives = 19/29 (64%)
Frame = +1
Query: 251 NEAFLMKILWNLIKNPEDLWCRVLRSKYG 279
N + L K W L+ + E LW RVL+++YG
Sbjct: 52 NLSLLGKWCWRLLVDKEGLWHRVLKARYG 138
Score = 28.1 bits (61), Expect(2) = 0.003
Identities = 10/26 (38%), Positives = 16/26 (61%)
Frame = +3
Query: 304 IWNDFKGHVVWNIGDGRNTNFWLDKW 329
+ N F+ ++ +GDGR+ FW D W
Sbjct: 228 VGNWFEENIRMVVGDGRDAFFWYDTW 305
>BG584442
Length = 775
Score = 38.9 bits (89), Expect = 0.007
Identities = 30/112 (26%), Positives = 49/112 (42%), Gaps = 1/112 (0%)
Frame = +1
Query: 160 KIQSKLNGWKQQCLTLAGRITLAKSVISTIPYYHMQYAKIPKTICDEIDKIQRGFIWGDS 219
K K+N + +CL+ + K + +I Y M + + DEI+KI F W
Sbjct: 370 KFDKKINF*RNKCLSKVM*EVMIKYALQSISSYVMSIFLLLNSQVDEIEKIMNTFSWVHV 549
Query: 220 DQGRKA-HLISWDVCCLPKADGGLGFRQTHKMNEAFLMKILWNLIKNPEDLW 270
+ RK H +S + + K GG+GF N L K + + + N L+
Sbjct: 550 GENRKGMHWMS*EKLFVHKNYGGMGFTDFTTFNIPMLGKQV*SFLLNRTTLF 705
>BG646730
Length = 799
Score = 34.3 bits (77), Expect = 0.18
Identities = 12/35 (34%), Positives = 20/35 (56%)
Frame = +1
Query: 298 WKALVNIWNDFKGHVVWNIGDGRNTNFWLDKWVPN 332
W+++ NI + W+I +G+N W D W+PN
Sbjct: 409 WRSMFNIKDVIDLGSRWSISNGQNVRIWKDDWLPN 513
>TC81772
Length = 982
Score = 30.0 bits (66), Expect(2) = 0.42
Identities = 17/57 (29%), Positives = 27/57 (46%)
Frame = -2
Query: 274 LRSKYGRNNDLIASINAHPYDSPLWKALVNIWNDFKGHVVWNIGDGRNTNFWLDKWV 330
LRSK +L+ + H S +W ++ + + W IGDG + N W D W+
Sbjct: 762 LRSKIFFTLELLGVVIGHN-PSYIWCSV*TLRMVLEEGHHWKIGDGSSINIWTDHWL 595
Score = 21.6 bits (44), Expect(2) = 0.42
Identities = 8/24 (33%), Positives = 13/24 (53%)
Frame = -1
Query: 243 GFRQTHKMNEAFLMKILWNLIKNP 266
GF + ++ L K +W L+ NP
Sbjct: 853 GFHDIYGLDLVMLGKQVWKLLTNP 782
>TC85426 similar to SP|O23758|NLTP_CICAR Nonspecific lipid-transfer protein
precursor (LTP). [Chickpea Garbanzo] {Cicer arietinum},
complete
Length = 696
Score = 31.2 bits (69), Expect = 1.5
Identities = 20/46 (43%), Positives = 26/46 (56%)
Frame = +2
Query: 607 WGLLLDVAAFFVIQTVDGSKATLRRLELVMPYMLRCGDYIWVWTWL 652
W L+L VA F+ QTV S L L ++ Y L G+ WV+TWL
Sbjct: 512 WRLILYVALSFLYQTVFDS---LLPLHVLFLYEL-LGEIFWVFTWL 637
>AW980456
Length = 779
Score = 29.3 bits (64), Expect = 5.9
Identities = 11/42 (26%), Positives = 20/42 (47%)
Frame = -2
Query: 399 NTHHFTVQSAYSLQHQDCPILEGDWKSIWKWHGPHRIQTFIW 440
+ + F V+ + + P G+W IWK P ++Q +W
Sbjct: 160 SAYRFCVEELFDSSYLHRP---GNWSGIWKLKVPPKVQNLVW 44
>TC84229 weakly similar to GP|9294175|dbj|BAB02077.1 cytochrome p450
{Arabidopsis thaliana}, partial (20%)
Length = 836
Score = 29.3 bits (64), Expect = 5.9
Identities = 12/26 (46%), Positives = 16/26 (61%)
Frame = +3
Query: 305 WNDFKGHVVWNIGDGRNTNFWLDKWV 330
W DF+GHV + + +F LDKWV
Sbjct: 747 WIDFQGHVGFMKKTAKEMDFILDKWV 824
>TC82284 weakly similar to GP|11119510|gb|AAF37724.2 diaphanous protein
{Entamoeba histolytica}, partial (4%)
Length = 671
Score = 25.8 bits (55), Expect(2) = 6.6
Identities = 10/33 (30%), Positives = 16/33 (48%)
Frame = -3
Query: 398 TNTHHFTVQSAYSLQHQDCPILEGDWKSIWKWH 430
+NTH T + + L WK++W+WH
Sbjct: 345 SNTHQHTALALHLLA----------WKTLWRWH 277
Score = 21.6 bits (44), Expect(2) = 6.6
Identities = 6/9 (66%), Positives = 7/9 (77%)
Frame = -3
Query: 427 WKWHGPHRI 435
W+W GP RI
Sbjct: 168 WRWRGPWRI 142
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.338 0.149 0.503
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,241,296
Number of Sequences: 36976
Number of extensions: 431335
Number of successful extensions: 3278
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 1802
Number of HSP's successfully gapped in prelim test: 162
Number of HSP's that attempted gapping in prelim test: 1401
Number of HSP's gapped (non-prelim): 2074
length of query: 755
length of database: 9,014,727
effective HSP length: 103
effective length of query: 652
effective length of database: 5,206,199
effective search space: 3394441748
effective search space used: 3394441748
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146757.4


