

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146756.7 - phase: 0
(96 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
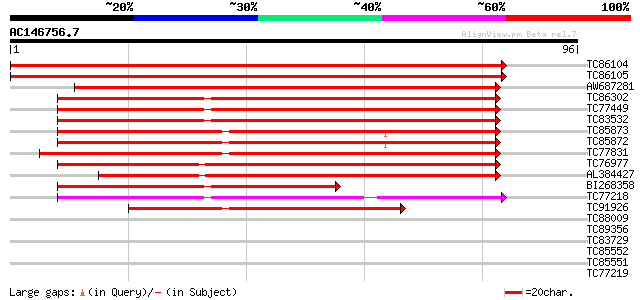
Score E
Sequences producing significant alignments: (bits) Value
TC86104 homologue to SP|O48551|PSA6_SOYBN Proteasome subunit alp... 170 1e-43
TC86105 homologue to SP|O48551|PSA6_SOYBN Proteasome subunit alp... 163 1e-41
AW687281 weakly similar to SP|Q9LSU3|PSA6 Proteasome subunit alp... 110 9e-26
TC86302 homologue to SP|Q9SXU1|PSA7_CICAR Proteasome subunit alp... 65 3e-12
TC77449 homologue to SP|Q9M4T8|PSA5_SOYBN Proteasome subunit alp... 65 3e-12
TC83532 similar to PIR|T38639|T38639 proteasome component PUP2 h... 65 4e-12
TC85873 homologue to SP|P52427|PSA4_SPIOL Proteasome subunit alp... 64 1e-11
TC85872 homologue to SP|P52427|PSA4_SPIOL Proteasome subunit alp... 64 1e-11
TC77831 homologue to GP|14594925|emb|CAC43323. putative alpha7 p... 63 2e-11
TC76977 homologue to SP|O23708|PSA2_ARATH Proteasome subunit alp... 60 2e-10
AL384427 similar to GP|18376026|em probable 20S proteasome subun... 57 1e-09
BI268358 homologue to GP|16769466|gb| LD33318p {Drosophila melan... 52 4e-08
TC77218 similar to GP|22947842|gb|AAN07899.1 20S proteasome alph... 50 2e-07
TC91926 similar to SP|P52427|PSA4_SPIOL Proteasome subunit alpha... 47 1e-06
TC88009 similar to PIR|T48507|T48507 probable GTP-binding protei... 28 0.77
TC89356 similar to GP|19911207|dbj|BAB86930. glucosyltransferase... 27 1.0
TC83729 similar to PIR|T39676|T39676 probable yeast cell divisio... 27 1.3
TC85552 similar to GP|17224608|gb|AAL37041.1 3-hydroxy-3-methylg... 26 2.9
TC85551 similar to GP|17224608|gb|AAL37041.1 3-hydroxy-3-methylg... 26 2.9
TC77219 similar to GP|22947842|gb|AAN07899.1 20S proteasome alph... 26 2.9
>TC86104 homologue to SP|O48551|PSA6_SOYBN Proteasome subunit alpha type 6
(EC 3.4.25.1) (20S proteasome alpha subunit A), complete
Length = 1113
Score = 170 bits (430), Expect = 1e-43
Identities = 84/84 (100%), Positives = 84/84 (100%)
Frame = +3
Query: 1 MSRGSGGGYDRHITIFSPEGRLFQVEYAFKAVKAAGITSIGVRGNDSVCVVTQKKVPDKL 60
MSRGSGGGYDRHITIFSPEGRLFQVEYAFKAVKAAGITSIGVRGNDSVCVVTQKKVPDKL
Sbjct: 150 MSRGSGGGYDRHITIFSPEGRLFQVEYAFKAVKAAGITSIGVRGNDSVCVVTQKKVPDKL 329
Query: 61 LDQTSVTHLFPITKYLGLLATGMT 84
LDQTSVTHLFPITKYLGLLATGMT
Sbjct: 330 LDQTSVTHLFPITKYLGLLATGMT 401
>TC86105 homologue to SP|O48551|PSA6_SOYBN Proteasome subunit alpha type 6
(EC 3.4.25.1) (20S proteasome alpha subunit A), complete
Length = 1047
Score = 163 bits (412), Expect = 1e-41
Identities = 80/84 (95%), Positives = 82/84 (97%)
Frame = +2
Query: 1 MSRGSGGGYDRHITIFSPEGRLFQVEYAFKAVKAAGITSIGVRGNDSVCVVTQKKVPDKL 60
MSRGSGGGYDRHITIFSPEGRLFQVEYAFKAVK+AGITSIGVRG DSVCVVTQKKVPDKL
Sbjct: 131 MSRGSGGGYDRHITIFSPEGRLFQVEYAFKAVKSAGITSIGVRGKDSVCVVTQKKVPDKL 310
Query: 61 LDQTSVTHLFPITKYLGLLATGMT 84
LD TSVTHLFPITKYLGLLATG+T
Sbjct: 311 LDHTSVTHLFPITKYLGLLATGIT 382
>AW687281 weakly similar to SP|Q9LSU3|PSA6 Proteasome subunit alpha type 6
(EC 3.4.25.1) (20S proteasome alpha subunit A), partial
(65%)
Length = 612
Score = 110 bits (275), Expect = 9e-26
Identities = 52/72 (72%), Positives = 62/72 (85%)
Frame = +3
Query: 12 HITIFSPEGRLFQVEYAFKAVKAAGITSIGVRGNDSVCVVTQKKVPDKLLDQTSVTHLFP 71
HITIFSPEGRL+QVEYAFKAVK G+TS+GVRG D V VV+QKKVPDKLLD TSVTHL+
Sbjct: 3 HITIFSPEGRLYQVEYAFKAVKEGGLTSVGVRGTDCVVVVSQKKVPDKLLDPTSVTHLYK 182
Query: 72 ITKYLGLLATGM 83
I+ ++G + TG+
Sbjct: 183ISDHIGAVVTGV 218
>TC86302 homologue to SP|Q9SXU1|PSA7_CICAR Proteasome subunit alpha type 7
(EC 3.4.25.1) (20S proteasome alpha subunit D), complete
Length = 1326
Score = 65.5 bits (158), Expect = 3e-12
Identities = 34/75 (45%), Positives = 48/75 (63%)
Frame = +2
Query: 9 YDRHITIFSPEGRLFQVEYAFKAVKAAGITSIGVRGNDSVCVVTQKKVPDKLLDQTSVTH 68
YDR IT+FSP+G LFQVEYA +AV+ G ++GVRG D+V + +KK KL D SV
Sbjct: 332 YDRAITVFSPDGHLFQVEYALEAVR-KGNAAVGVRGIDNVVLGVEKKSTAKLQDSRSVRK 508
Query: 69 LFPITKYLGLLATGM 83
+ + ++ L G+
Sbjct: 509 IVNLDDHIALACAGL 553
>TC77449 homologue to SP|Q9M4T8|PSA5_SOYBN Proteasome subunit alpha type 5
(EC 3.4.25.1) (20S proteasome alpha subunit E), complete
Length = 1091
Score = 65.5 bits (158), Expect = 3e-12
Identities = 31/75 (41%), Positives = 49/75 (65%)
Frame = +2
Query: 9 YDRHITIFSPEGRLFQVEYAFKAVKAAGITSIGVRGNDSVCVVTQKKVPDKLLDQTSVTH 68
YDR + FSPEGRLFQVEYA +A+K G T+IG++ + V + +K++ LL+ +SV
Sbjct: 119 YDRGVNTFSPEGRLFQVEYAIEAIK-LGSTAIGLKTKEGVVLAVEKRITSPLLEPSSVEK 295
Query: 69 LFPITKYLGLLATGM 83
+ I ++G +G+
Sbjct: 296 IMEIDDHIGCAMSGL 340
>TC83532 similar to PIR|T38639|T38639 proteasome component PUP2 homolog -
fission yeast (Schizosaccharomyces pombe), partial (97%)
Length = 901
Score = 65.1 bits (157), Expect = 4e-12
Identities = 33/75 (44%), Positives = 50/75 (66%)
Frame = +3
Query: 9 YDRHITIFSPEGRLFQVEYAFKAVKAAGITSIGVRGNDSVCVVTQKKVPDKLLDQTSVTH 68
YDR I FSPEGRLFQVEY+ +A+K G T+IGV ++ V + +K+V LL+ +SV
Sbjct: 96 YDRGINTFSPEGRLFQVEYSLEAIK-LGSTAIGVATSEGVVLGVEKRVTSTLLETSSVEK 272
Query: 69 LFPITKYLGLLATGM 83
+ I +++G +G+
Sbjct: 273 IVEIDRHIGCAMSGL 317
>TC85873 homologue to SP|P52427|PSA4_SPIOL Proteasome subunit alpha type 4
(EC 3.4.25.1) (20S proteasome alpha subunit C), partial
(98%)
Length = 1042
Score = 63.5 bits (153), Expect = 1e-11
Identities = 35/76 (46%), Positives = 47/76 (61%), Gaps = 1/76 (1%)
Frame = +2
Query: 9 YDRHITIFSPEGRLFQVEYAFKAVKAAGITSIGVRGNDSVCVVTQKKVPDKLLD-QTSVT 67
YD TIFSPEGRL+QVEYA +A+ AG T+IG+ D V +V +KKV KLL TS
Sbjct: 83 YDSRTTIFSPEGRLYQVEYAMEAIGNAG-TAIGILSKDGVVLVGEKKVTSKLLQTSTSTE 259
Query: 68 HLFPITKYLGLLATGM 83
++ I ++ G+
Sbjct: 260 KMYKIDDHVACAVAGI 307
>TC85872 homologue to SP|P52427|PSA4_SPIOL Proteasome subunit alpha type 4
(EC 3.4.25.1) (20S proteasome alpha subunit C), complete
Length = 1139
Score = 63.5 bits (153), Expect = 1e-11
Identities = 35/76 (46%), Positives = 47/76 (61%), Gaps = 1/76 (1%)
Frame = +2
Query: 9 YDRHITIFSPEGRLFQVEYAFKAVKAAGITSIGVRGNDSVCVVTQKKVPDKLLD-QTSVT 67
YD TIFSPEGRL+QVEYA +A+ AG T+IG+ D V +V +KKV KLL TS
Sbjct: 140 YDSRTTIFSPEGRLYQVEYAMEAIGNAG-TAIGILSKDGVVLVGEKKVTSKLLQTSTSTE 316
Query: 68 HLFPITKYLGLLATGM 83
++ I ++ G+
Sbjct: 317 KMYKIDDHVACAVAGI 364
>TC77831 homologue to GP|14594925|emb|CAC43323. putative alpha7 proteasome
subunit {Nicotiana tabacum}, complete
Length = 1109
Score = 62.8 bits (151), Expect = 2e-11
Identities = 30/78 (38%), Positives = 48/78 (61%)
Frame = +1
Query: 6 GGGYDRHITIFSPEGRLFQVEYAFKAVKAAGITSIGVRGNDSVCVVTQKKVPDKLLDQTS 65
G GYD +T FSP+GR+FQ+EYA KAV +G T IG+R D + + +K +P K++ S
Sbjct: 91 GTGYDLSVTTFSPDGRVFQIEYAAKAVDNSG-TVIGIRCKDGIVLGVEKLIPSKMMLSGS 267
Query: 66 VTHLFPITKYLGLLATGM 83
+ + ++ G+ G+
Sbjct: 268 NRRIHAVHRHSGMAVAGL 321
>TC76977 homologue to SP|O23708|PSA2_ARATH Proteasome subunit alpha type 2
(EC 3.4.25.1) (20S proteasome alpha subunit B).
[Mouse-ear cress], complete
Length = 1150
Score = 59.7 bits (143), Expect = 2e-10
Identities = 29/75 (38%), Positives = 49/75 (64%)
Frame = +3
Query: 9 YDRHITIFSPEGRLFQVEYAFKAVKAAGITSIGVRGNDSVCVVTQKKVPDKLLDQTSVTH 68
Y +T FSP G+L Q+E+A AV +G TS+G++ + V + T+KK+P L+D+ SV
Sbjct: 120 YSFSLTTFSPSGKLVQIEHALTAV-GSGQTSLGIKAANGVVIATEKKLPSILVDEASVQK 296
Query: 69 LFPITKYLGLLATGM 83
+ +T +G++ +GM
Sbjct: 297 IQLLTPNIGVVYSGM 341
>AL384427 similar to GP|18376026|em probable 20S proteasome subunit Y7
{Neurospora crassa}, partial (59%)
Length = 462
Score = 57.0 bits (136), Expect = 1e-09
Identities = 26/68 (38%), Positives = 44/68 (64%)
Frame = +2
Query: 16 FSPEGRLFQVEYAFKAVKAAGITSIGVRGNDSVCVVTQKKVPDKLLDQTSVTHLFPITKY 75
FSP G+L Q+EYA AV A G+TSIG++ + + + T+KK L+D +S+ + +
Sbjct: 2 FSPSGKLVQIEYALNAV-AQGVTSIGIKATNGIVIATEKKSASPLIDDSSLEKVANVCPN 178
Query: 76 LGLLATGM 83
+G++ +GM
Sbjct: 179IGMVYSGM 202
>BI268358 homologue to GP|16769466|gb| LD33318p {Drosophila melanogaster},
partial (22%)
Length = 325
Score = 52.0 bits (123), Expect = 4e-08
Identities = 24/48 (50%), Positives = 35/48 (72%)
Frame = +1
Query: 9 YDRHITIFSPEGRLFQVEYAFKAVKAAGITSIGVRGNDSVCVVTQKKV 56
YDR + FSPEGRLFQVEYA +A+K G T+IG+ ++ V + +K++
Sbjct: 169 YDRGVNTFSPEGRLFQVEYAIEAIK-LGSTAIGICTSEGVVLAVEKRI 309
>TC77218 similar to GP|22947842|gb|AAN07899.1 20S proteasome alpha 6 subunit
{Nicotiana benthamiana}, partial (91%)
Length = 1236
Score = 49.7 bits (117), Expect = 2e-07
Identities = 28/76 (36%), Positives = 42/76 (54%)
Frame = +2
Query: 9 YDRHITIFSPEGRLFQVEYAFKAVKAAGITSIGVRGNDSVCVVTQKKVPDKLLDQTSVTH 68
YD +T +SP GRLFQVEYA +AVK G +IG+R V + K +L +
Sbjct: 86 YDTDVTTWSPAGRLFQVEYAMEAVK-QGSAAIGLRSKTHVVLACVNKANSEL--SSHQKK 256
Query: 69 LFPITKYLGLLATGMT 84
+F + ++G+ G+T
Sbjct: 257 IFKVDDHIGVAIAGLT 304
>TC91926 similar to SP|P52427|PSA4_SPIOL Proteasome subunit alpha type 4
(EC 3.4.25.1) (20S proteasome alpha subunit C), partial
(78%)
Length = 728
Score = 47.0 bits (110), Expect = 1e-06
Identities = 21/47 (44%), Positives = 34/47 (71%)
Frame = +1
Query: 21 RLFQVEYAFKAVKAAGITSIGVRGNDSVCVVTQKKVPDKLLDQTSVT 67
RL+QVEYA +A+ AG T++G+ +D + + +KKV KLL+QT+ +
Sbjct: 1 RLYQVEYAMEAISHAG-TALGILASDGIVIAAEKKVTSKLLEQTTTS 138
>TC88009 similar to PIR|T48507|T48507 probable GTP-binding protein -
Arabidopsis thaliana, partial (72%)
Length = 1399
Score = 27.7 bits (60), Expect = 0.77
Identities = 12/31 (38%), Positives = 18/31 (57%), Gaps = 4/31 (12%)
Frame = +2
Query: 45 NDSVCVVTQKKVP----DKLLDQTSVTHLFP 71
N S+C VTQ K P D++ +T + +FP
Sbjct: 146 NPSICEVTQAKTPT*ETDRIASKTLIAEIFP 238
>TC89356 similar to GP|19911207|dbj|BAB86930. glucosyltransferase-12 {Vigna
angularis}, partial (58%)
Length = 1642
Score = 27.3 bits (59), Expect = 1.0
Identities = 13/35 (37%), Positives = 21/35 (59%)
Frame = -3
Query: 60 LLDQTSVTHLFPITKYLGLLATGMTGVSFIASPSF 94
+ D T+ + LF IT + T ++ SFI++PSF
Sbjct: 1358 IADLTNSSFLFTITSFTIFSTTFLSQTSFISNPSF 1254
>TC83729 similar to PIR|T39676|T39676 probable yeast cell division cycle
CDC50 homolog - fission yeast (Schizosaccharomyces
pombe), partial (19%)
Length = 557
Score = 26.9 bits (58), Expect = 1.3
Identities = 12/34 (35%), Positives = 20/34 (58%)
Frame = +3
Query: 63 QTSVTHLFPITKYLGLLATGMTGVSFIASPSFLL 96
Q + +LFP+TKY G + ++ VSF+ + L
Sbjct: 183 QFEIQYLFPVTKYGGTKSLVISTVSFLGGKNSFL 284
>TC85552 similar to GP|17224608|gb|AAL37041.1 3-hydroxy-3-methylglutaryl
coenzyme A {Pisum sativum}, partial (81%)
Length = 2008
Score = 25.8 bits (55), Expect = 2.9
Identities = 17/53 (32%), Positives = 23/53 (43%)
Frame = +2
Query: 33 KAAGITSIGVRGNDSVCVVTQKKVPDKLLDQTSVTHLFPITKYLGLLATGMTG 85
KAA + I RG VC K+ K + +TSV L + L + M G
Sbjct: 1130 KAAAVNWIEGRGKSVVCEAVIKEEVVKKVLKTSVESLVELNMLKNLTGSAMAG 1288
>TC85551 similar to GP|17224608|gb|AAL37041.1 3-hydroxy-3-methylglutaryl
coenzyme A {Pisum sativum}, partial (83%)
Length = 2059
Score = 25.8 bits (55), Expect = 2.9
Identities = 17/53 (32%), Positives = 23/53 (43%)
Frame = +2
Query: 33 KAAGITSIGVRGNDSVCVVTQKKVPDKLLDQTSVTHLFPITKYLGLLATGMTG 85
KAA + I RG VC K+ K + +TSV L + L + M G
Sbjct: 1154 KAAAVNWIEGRGKSVVCEAVIKEEVVKKVLKTSVESLVELNMLKNLTGSAMAG 1312
>TC77219 similar to GP|22947842|gb|AAN07899.1 20S proteasome alpha 6 subunit
{Nicotiana benthamiana}, partial (22%)
Length = 692
Score = 25.8 bits (55), Expect = 2.9
Identities = 12/23 (52%), Positives = 14/23 (60%)
Frame = +2
Query: 9 YDRHITIFSPEGRLFQVEYAFKA 31
Y I +SP G FQVEYA +A
Sbjct: 476 YHTDIATWSPAGGHFQVEYAMEA 544
Score = 25.4 bits (54), Expect = 3.8
Identities = 11/23 (47%), Positives = 14/23 (60%)
Frame = +2
Query: 9 YDRHITIFSPEGRLFQVEYAFKA 31
Y + +SP G FQVEYA +A
Sbjct: 245 YHTDVATWSPAGGHFQVEYAMEA 313
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.139 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,363,235
Number of Sequences: 36976
Number of extensions: 23316
Number of successful extensions: 107
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 103
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 104
length of query: 96
length of database: 9,014,727
effective HSP length: 72
effective length of query: 24
effective length of database: 6,352,455
effective search space: 152458920
effective search space used: 152458920
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 50 (23.9 bits)
Medicago: description of AC146756.7


