

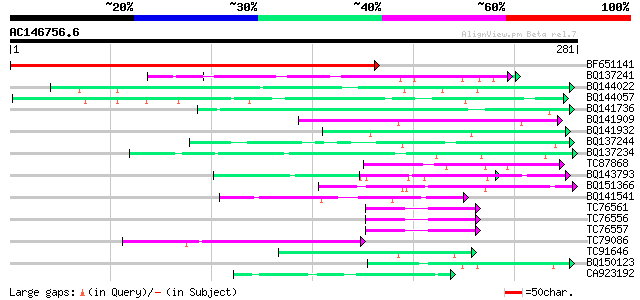
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146756.6 - phase: 0
(281 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BF651141 homologue to GP|387018|gb|AA prostaglandin-endoperoxide... 317 3e-87
BQ137241 similar to GP|6523547|emb| hydroxyproline-rich glycopro... 56 1e-08
BQ144022 homologue to GP|22831315|dbj hypothetical protein~predi... 56 1e-08
BQ144057 similar to GP|1483567|emb| viral proteinase {Pseudorabi... 52 3e-07
BQ141736 similar to PIR|T06291|T062 extensin homolog T9E8.80 - A... 51 4e-07
BQ141909 weakly similar to GP|14253159|emb VMP3 protein {Volvox ... 48 5e-06
BQ141932 weakly similar to GP|6523547|emb hydroxyproline-rich gl... 47 1e-05
BQ137244 similar to GP|20161222|dbj Epstein-Barr virus EBNA-1-li... 47 1e-05
BQ137234 similar to EGAD|35719|3709 hypothetical protein {Burkho... 45 3e-05
TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR... 44 5e-05
BQ143793 weakly similar to GP|6523547|emb| hydroxyproline-rich g... 44 7e-05
BQ151366 similar to GP|20146300|dbj hypothetical protein~similar... 44 7e-05
BQ141541 similar to GP|334068|gb|A ORF2 {Pseudorabies virus}, pa... 44 9e-05
TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 43 1e-04
TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 43 1e-04
TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 43 1e-04
TC79086 similar to GP|23499033|emb|CAD51113. ubiquitin-protein l... 43 2e-04
TC91646 weakly similar to GP|6523547|emb|CAB62280.1 hydroxyproli... 42 3e-04
BQ150123 similar to GP|21109992|gb| TonB-dependent receptor {Xan... 40 8e-04
CA923192 similar to GP|20196906|gb| expressed protein {Arabidops... 40 8e-04
>BF651141 homologue to GP|387018|gb|AA prostaglandin-endoperoxide synthase-1
{Homo sapiens}, partial (2%)
Length = 603
Score = 317 bits (812), Expect = 3e-87
Identities = 160/183 (87%), Positives = 163/183 (88%)
Frame = +1
Query: 1 MKTFGQAQSNTITQKSDSLPPNTPLSRPDSLSKHKPPILVCRKRSSIPVKPSTTTFSHKG 60
MKTFGQAQSNTITQKSDSLPPNTPLSRPDSLSKHKPPILVCRKRSSIPVKPSTTTFSHKG
Sbjct: 55 MKTFGQAQSNTITQKSDSLPPNTPLSRPDSLSKHKPPILVCRKRSSIPVKPSTTTFSHKG 234
Query: 61 DHKPIINDKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNK 120
DHKPIINDKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNK
Sbjct: 235 DHKPIINDKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNK 414
Query: 121 KPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSG 180
KPPSNSTPRTGSSANKPAETPKLKNKAEGVSDK++ FL K+ G
Sbjct: 415 KPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKEEXQRGCWFLEXNKEK*IGGXAEXWYG 594
Query: 181 GGG 183
GGG
Sbjct: 595 GGG 603
>BQ137241 similar to GP|6523547|emb| hydroxyproline-rich glycoprotein DZ-HRGP
{Volvox carteri f. nagariensis}, partial (8%)
Length = 1176
Score = 56.2 bits (134), Expect = 1e-08
Identities = 53/196 (27%), Positives = 81/196 (41%), Gaps = 15/196 (7%)
Frame = +1
Query: 69 KKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNKKPPSNSTP 128
KK+R D T PS T + PP+ GA+ RR A + T
Sbjct: 52 KKKRSEGDQARTF-PSLKGTKDGAPPEG-----AGAQKRRGGHKAGGKAPRGRAQKKRTT 213
Query: 129 RTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGS 188
+TGS AE P N + + ++N G+ + KKN + R +G
Sbjct: 214 KTGS-----AEKPPQPNHPKQPTATERNTGS-----QAKKNTKRKKTRPTTGDADAQQKR 363
Query: 189 SKGGGGGGTNT---KEQKKVVNNGKGEKGKERASRYND---GGGSGSGD----KRNKNIE 238
++GG G T T K Q++ K E+G+++ R + GGG G+G+ +RN+ E
Sbjct: 364 AEGGSGRETRTHAQKTQEQTARREKREEGRKKRKRAREGEKGGGEGAGNRAEQRRNEQRE 543
Query: 239 -----SNSKRNVGRPP 249
+ R GR P
Sbjct: 544 EWHTHTKETRGNGRKP 591
Score = 46.2 bits (108), Expect = 1e-05
Identities = 39/163 (23%), Positives = 66/163 (39%), Gaps = 6/163 (3%)
Frame = +3
Query: 97 KEKPVTGARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKN 156
K + TG R K + + + P ST + K ETP + D+KK+
Sbjct: 108 KRRSTTGGGGRPKTKG-RAQSRRESAPRESTEKKNHQNRKRGETPTTEPPETADRDRKKH 284
Query: 157 NGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGG------GGGGTNTKEQKKVVNNGK 210
++ K+NE + GG ++ S+GG +T+ K G+
Sbjct: 285 GQPGQKKHQTKENE------ADDGGRRRTTKKSRGGERTRNPNARTKDTRTNSKAGEEGR 446
Query: 211 GEKGKERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKAA 253
EK +E ++R GGG G G K+++ + R V ++ A
Sbjct: 447 REKEEEESARGRKGGGRG-GRKQSRAATQRTTRRVAHTHERDA 572
>BQ144022 homologue to GP|22831315|dbj hypothetical protein~predicted by
GlimmerM etc. {Oryza sativa (japonica cultivar-group)},
partial (16%)
Length = 1286
Score = 56.2 bits (134), Expect = 1e-08
Identities = 64/290 (22%), Positives = 104/290 (35%), Gaps = 30/290 (10%)
Frame = +2
Query: 21 PNTPLSRPDSLSK-----HKPPILVCRKRSSIPVKPS---TTTFSHKGDHKPIINDKKER 72
PN P +P + + H P KR KP+ T + K +P +
Sbjct: 17 PNAPAPQPHTETTQRFLGHTPQNFSSLKRKHKTSKPTHERRTARNKKQRGQPTHTRPRTP 196
Query: 73 PSSDVKPTLKPSYSETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNKKPPSNSTPRTGS 132
S +P+ ++ P+A+ P R+R + S+ +N + P + PR
Sbjct: 197 TPSRAQPSSATKTDRRPDDARPEARPHPRGQTRNRPDPEPNESHRTNSRTPE-TIPRHSP 373
Query: 133 SANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGG 192
+ +P + + S K K G + +G GGGGG G +GG
Sbjct: 374 AQTQPGSDLSRQETNQYPSTKPKKADGGGVAGP-------K*PTAGEGGGGGGGGEGEGG 532
Query: 193 GG-----------GGTNTKEQKKVVNNGKGEK--GKERASRYNDGGGSGSG--------- 230
G GG ++ + G+G + GKER GG G G
Sbjct: 533 EGRAGKREEKK*KGGPPL*TERAGGDKGRGARKGGKERVQHAKPGGAGGEGRVCRHRNRP 712
Query: 231 DKRNKNIESNSKRNVGRPPKKAAETVASTKRGRESSASGGKDKRPKKRSK 280
++ NI+S R R + T T+R R + K R ++R +
Sbjct: 713 EREEGNIKSERAREREREARAEGTTDEWTRRERRARTRTSKRGRGRQRER 862
>BQ144057 similar to GP|1483567|emb| viral proteinase {Pseudorabies virus},
partial (5%)
Length = 1295
Score = 51.6 bits (122), Expect = 3e-07
Identities = 66/295 (22%), Positives = 108/295 (36%), Gaps = 19/295 (6%)
Frame = +1
Query: 2 KTFGQAQSNTITQKSDSLPPNTPLSRPDSLSKHKP-PILVCRKRSSIPVKPS------TT 54
K F + ++ T S+ P P+ P +L+ + P P R S P +P+ T
Sbjct: 88 KNFSRVKTKTREGTSNETPTRNPIHTPTTLTPNPPAPREAARPAPSHPARPTRRIPHRTP 267
Query: 55 TFSHKGDHKPII-----NDKKERPSSDVKPTLK-PSYSETDEEEPPKAKEKPVTGARSRR 108
FS H+PI +K P + TL P+ + + P++ R+RR
Sbjct: 268 QFSSTHTHQPIPYKTHQTNKTSNPRTQQTTTLSTPTPTPSPPRTRPRSPNAQTPSDRTRR 447
Query: 109 SNKNLSSNA--SNKKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRI 166
+ +L + A + P P+T P +PK K G +
Sbjct: 448 AG-HLQAGAFPGRAQGPGP*QPQT------PGASPKTK*MRGGGAKSVDGRDRGRRERAS 606
Query: 167 KKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDG-- 224
++N + G+GGGG S +G G T++ KGE+ + +R G
Sbjct: 607 RENAGARRAQRGAGGGGPRS---RGAHRRGNGTRQ--------KGERKRRAEARGQQGTS 753
Query: 225 --GGSGSGDKRNKNIESNSKRNVGRPPKKAAETVASTKRGRESSASGGKDKRPKK 277
G+G R E R RP + A RG G +++ P +
Sbjct: 754 EERSGGAGRSRGTASEMQCARGTTRPRRATAHW-----RGGREDERGAREETPTR 903
>BQ141736 similar to PIR|T06291|T062 extensin homolog T9E8.80 - Arabidopsis
thaliana, partial (4%)
Length = 1099
Score = 51.2 bits (121), Expect = 4e-07
Identities = 40/188 (21%), Positives = 71/188 (37%), Gaps = 1/188 (0%)
Frame = +2
Query: 94 PKAKEKPVTGARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDK 153
P+AK++ G + RR + +N + +T R G K ++ +K
Sbjct: 134 PEAKKEHARG-QPRRGPRPTYNNPRPTNTQNRNTRRRGKRKEKKRSLRPPRHAPPARPEK 310
Query: 154 KKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEK 213
GAAG + + R + GGG+ + G GT T ++K +
Sbjct: 311 AGPGGAAGGGGGDRTTRKKDRRRQRTAAGGGTGAGKEAATGAGTKTGGRRKTTGGDRPTP 490
Query: 214 GKERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKAAETVASTKRGRESS-ASGGKD 272
G+E+ ++ G+ N K +K + T+ GR GG++
Sbjct: 491 GREKKNQRTKESGA--------RAPKNQKERAAGGRRKGGKEAGGTETGRSGEREGGGRE 646
Query: 273 KRPKKRSK 280
+R KKR +
Sbjct: 647 ERKKKRKR 670
Score = 35.4 bits (80), Expect = 0.025
Identities = 44/234 (18%), Positives = 82/234 (34%), Gaps = 1/234 (0%)
Frame = +1
Query: 49 VKPSTTTFSHKGDHKPIINDKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVTGARSRR 108
VK +H+G + N+K+ P D + + + P +P + +
Sbjct: 37 VKRDKAGDTHRGGGRN--NNKRREP--DTNGHRRQASGSQERTRPRPTPPRPTADLQQPQ 204
Query: 109 SNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKK 168
++K+ ++K + + +PA + G + + G R +
Sbjct: 205 ADKHTKPKHASKGETKGKEKKFTAPQTRPARPAREGGPRRG--GRGRGRGPDDPQERQTE 378
Query: 169 NESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSG 228
E+ GG G +G KGG GG + + N + + R + + G
Sbjct: 379 TEN---------GGRGGNGGRKGGSDGGGDENGGAEKDNRRRPADPRPRKEKPTNKRKRG 531
Query: 229 SGDKRNKNIESNSKRNVGRPPKKAAET-VASTKRGRESSASGGKDKRPKKRSKK 281
+G K+ KR R KK E R + GGK + K++ K+
Sbjct: 532 TGTKK-------PKREGRRGEKKGRERGRGDGNREKRGERGGGKGRTKKEKEKR 672
>BQ141909 weakly similar to GP|14253159|emb VMP3 protein {Volvox carteri f.
nagariensis}, partial (13%)
Length = 1223
Score = 47.8 bits (112), Expect = 5e-06
Identities = 36/140 (25%), Positives = 57/140 (40%), Gaps = 9/140 (6%)
Frame = +2
Query: 144 KNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKG-----GGGGGTN 198
+NK G + K+ G ++ E + G GGGG GS +G GGG G
Sbjct: 95 ENKNNGRNTTKEGRRGRGSGKMRRRGERGKRGEGGEGGGGAEGGSERGRGRRGGGGRGGG 274
Query: 199 TKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKA----AE 254
+ + K G +GK +GGG G + RN + G+ +K+ +
Sbjct: 275 RRGRGKKGGGGTKGRGKRERGGGTEGGGKGKIEGRNGTGRRRGESGGGKGREKSELLQTD 454
Query: 255 TVASTKRGRESSASGGKDKR 274
+S + S GGK++R
Sbjct: 455 VESSLQELTHSIWEGGKERR 514
Score = 36.6 bits (83), Expect = 0.011
Identities = 30/106 (28%), Positives = 43/106 (40%), Gaps = 2/106 (1%)
Frame = +1
Query: 178 GSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKRNKNI 237
G GGGG G GGG G K ++ G+GE+G +G G +KR
Sbjct: 178 GEAGGGGGGGGGSGGGVG--EGKREEGGGGEGRGEEG--------EGEERGGRNKRKGEA 327
Query: 238 ESNSKRNVGRPPKKAAETVASTKRGRE--SSASGGKDKRPKKRSKK 281
+ GR K RG+E +GGK + ++R K+
Sbjct: 328 *ERGRNGGGRQGK---------NRGKEWDREEAGGKWRGKREREKR 438
Score = 36.2 bits (82), Expect = 0.014
Identities = 36/131 (27%), Positives = 56/131 (42%), Gaps = 14/131 (10%)
Frame = +2
Query: 164 NRIKKNESVEVLRSGSGGG------GGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKER 217
+R + ES +R G G GGS G++ G G GG+ E + V + +K +ER
Sbjct: 821 DRYHERESR*GVRGGERRGARREEEGGSEGAATGNGAGGS---EA*RGVEWSE*QKCEER 991
Query: 218 ASRYNDG----GGSGSGDKRNKNIESNSKRNVGR----PPKKAAETVASTKRGRESSASG 269
R G G SG G+ R E +R R KK ++ GR + G
Sbjct: 992 EVRERWGGERTGESGGGE*RQSERERRRERRRTRKQIAEEKK*GGREGRSETGRRRAGGG 1171
Query: 270 GKDKRPKKRSK 280
G+++ ++ K
Sbjct: 1172GREREQRREMK 1204
Score = 35.0 bits (79), Expect = 0.032
Identities = 31/118 (26%), Positives = 42/118 (35%)
Frame = +3
Query: 163 LNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYN 222
+ R K+ EV+R G GGS G +GG GGG G +G+E
Sbjct: 117 IQRRKEGGGEEVVR*EEGVKGGSGG--RGGRGGGERRG----------GRRGEEGGGGGG 260
Query: 223 DGGGSGSGDKRNKNIESNSKRNVGRPPKKAAETVASTKRGRESSASGGKDKRPKKRSK 280
G G G G R E +V ++ + G GGK + K K
Sbjct: 261 GGEGGGGGGGRKGGEEQKEGGSVREGEERRGAAREK*REGMGQGGGGGKVEGEKGERK 434
Score = 29.6 bits (65), Expect = 1.4
Identities = 39/187 (20%), Positives = 61/187 (31%), Gaps = 25/187 (13%)
Frame = +3
Query: 103 GARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGA--A 160
G R + + NA N+ T S A P K + EG K +GA A
Sbjct: 618 GTVKRNAERGEDGNARNEGKEERMRIGTISIAAGPIRQEK---RREGRK*NAKESGARSA 788
Query: 161 GFLNRIKKNESVEVLRSGSGG-----------------------GGGSSGSSKGGGGGGT 197
L R + +++++R +G G G+ G
Sbjct: 789 YRLGRDRGKRAIDIMRGRAGEE*EGERDEERDGRRREGARARRRGTAQEGARHDAESSGA 968
Query: 198 NTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKAAETVA 257
N + V + + E+G R GGGS K N + + + + K E
Sbjct: 969 NNRS----VRSAR*ERGGAARGRERAGGGSRGNQKGNAGGNGDERESR*QKRKNEGEEKG 1136
Query: 258 STKRGRE 264
+RG E
Sbjct: 1137GARRGGE 1157
Score = 28.5 bits (62), Expect = 3.0
Identities = 20/73 (27%), Positives = 29/73 (39%), Gaps = 7/73 (9%)
Frame = +2
Query: 182 GGGSSGSSKGGGGGGTNTKEQKKVVNNGKG-----EKGKERASRYNDGGGSGSGDKRN-- 234
GGG +G GGG + + K +G E+G+ R + Y S DK
Sbjct: 560 GGGRAGDRGGGGLRAPSKRRNSKTKRRKRGGRECPERGEGRTNAYRHNIDSSGADKAREA 739
Query: 235 KNIESNSKRNVGR 247
+ E + VGR
Sbjct: 740 ERGEEVKRERVGR 778
>BQ141932 weakly similar to GP|6523547|emb hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (30%)
Length = 1338
Score = 46.6 bits (109), Expect = 1e-05
Identities = 37/137 (27%), Positives = 50/137 (36%), Gaps = 14/137 (10%)
Frame = +3
Query: 156 NNGAAGFLNRIKKNESVEVLRS-------------GSGGGGGSSGSSKGGGGGGTNTKEQ 202
N GA FLN IK + +L G+GGGGG G +G G G +E+
Sbjct: 54 NKGAYEFLNMIKTSSYFYLLSGVYPAAGA*IGVVVGAGGGGGGVGGGRGRGRAG*GEEER 233
Query: 203 KKVVNNGKGEKGKERASRYNDGGGS-GSGDKRNKNIESNSKRNVGRPPKKAAETVASTKR 261
+ V + E G + GGG G+ RN + + TV
Sbjct: 234 GRGVGGERRELGGRMEGEWGGGGGGRRGGESRNGYKGRRGSGTADAAGGRGSMTVGDRGG 413
Query: 262 GRESSASGGKDKRPKKR 278
G S G + R K R
Sbjct: 414 GGRSEGGGAEGGRAKAR 464
Score = 29.3 bits (64), Expect = 1.8
Identities = 28/93 (30%), Positives = 39/93 (41%), Gaps = 5/93 (5%)
Frame = +2
Query: 141 PKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTK 200
P+ +AEG + + GA +R + EV G GGGGG S ++G GGG
Sbjct: 326 PQWVQRAEGEWNCRCGGGAR-VDDRGRPGRGGEV--GGWGGGGGESEGARGWGGG----- 481
Query: 201 EQKKVVNNGKGEKGKERASRYN-----DGGGSG 228
+G G+ G R+ D GG G
Sbjct: 482 ------RDGGGQAGVWLVGRFRVAGDWDAGGLG 562
Score = 26.9 bits (58), Expect = 8.8
Identities = 19/70 (27%), Positives = 26/70 (37%), Gaps = 26/70 (37%)
Frame = +1
Query: 177 SGSGGGGGSSGSSKGG--------------------------GGGGTNTKEQKKVVNNGK 210
+G+GGGG G +GG GGGG + VV+ G
Sbjct: 403 TGAGGGGRRVGGRRGGERRRAWVGGRTGWGGAGRGVAGGSVQGGGGLGCWGVRVVVDGGW 582
Query: 211 GEKGKERASR 220
G G ++ SR
Sbjct: 583 GAAGSQKRSR 612
>BQ137244 similar to GP|20161222|dbj Epstein-Barr virus EBNA-1-like protein
{Oryza sativa (japonica cultivar-group)}, partial (5%)
Length = 1050
Score = 46.6 bits (109), Expect = 1e-05
Identities = 53/205 (25%), Positives = 71/205 (33%), Gaps = 14/205 (6%)
Frame = +2
Query: 90 EEEPPKAKEKPVTGARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNKAEG 149
E +PP E G R SN + PP+N+TP TG A A P
Sbjct: 2 ERDPPGGSECGAEGGRRDESN-------NKPHPPTNATPETGQDATARASHPTRSR---- 148
Query: 150 VSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGG-------------GGG 196
D K N RIKK GGG+ G KG G G
Sbjct: 149 --DNKYN------*RRIKKY------------GGGNDGEKKGRGEEPPKPS**KQKNEGR 268
Query: 197 TNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKAAETV 256
T K + V K E G E + G G K + ++ R R +T
Sbjct: 269 T*EKTGPQAVGRNKKETGTE-----TETGPRGGQRKEREQAPAHGHRRAARKAANTRDT* 433
Query: 257 ASTKRGRESSASG-GKDKRPKKRSK 280
R R+S+ +G G+D P + ++
Sbjct: 434 IHDVRPRDSARAGAGRDAVPARETR 508
>BQ137234 similar to EGAD|35719|3709 hypothetical protein {Burkholderia
cepacia}, partial (6%)
Length = 1184
Score = 45.1 bits (105), Expect = 3e-05
Identities = 55/247 (22%), Positives = 91/247 (36%), Gaps = 25/247 (10%)
Frame = +1
Query: 60 GDHKPIINDKKERPSSDVKPTLKPSYSETDEEEPPKAKEKPVTGARSRRSNKNLSSNASN 119
G +K +KER ++ KP K + EE K K++P R R K A
Sbjct: 253 GRNKRRSQRQKERKENERKPKTK---QKKGGEETRKKKKQPQR-RRGEREEKGEGEEARK 420
Query: 120 KKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGS 179
++ P + + +K E K+K + S +N R E E R S
Sbjct: 421 REGPGRTNRKQKRRTHK--EESGKKSKQQQRSTSTAHNTQKRETTRAPARERAETARVTS 594
Query: 180 GGGGGSSGSSKGGGGGGTNTKEQKKVVNNGK------------------GEKGKERASRY 221
G S+G++K K +++ G+ GE+G+ER +
Sbjct: 595 NAGDDSTGAAKR-----IREKTRRRAAERGRDTPRHDGQPTHREARRHVGERGEERDNET 759
Query: 222 NDGGGSGSGDK----RNKNIESNSKRNVGRPPKKAAETVASTKRGRE---SSASGGKDKR 274
+ DK R+KN ++ P +++ T A ++ E A G DK
Sbjct: 760 EGSRAATDADKKTRRRSKNTGRENRAREAAPARESERTRAQHEKHAEQRGEDARGRTDKS 939
Query: 275 PKKRSKK 281
+ SK+
Sbjct: 940 ARTSSKR 960
>TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana, partial (91%)
Length = 860
Score = 44.3 bits (103), Expect = 5e-05
Identities = 33/108 (30%), Positives = 49/108 (44%), Gaps = 8/108 (7%)
Frame = +3
Query: 176 RSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGK-ERASRYNDGGGSGSGDKRN 234
RSG GGGGG G +GGGGGG + + + GE G R R GGG+G R+
Sbjct: 258 RSGGGGGGGGGGRGRGGGGGGGSDLKCYEC-----GEPGHFARECRNRGGGGAGRRRSRS 422
Query: 235 ----KNIESNSKRNV---GRPPKKAAETVASTKRGRESSASGGKDKRP 275
+ S +R+ GR P++ + + R G+++ P
Sbjct: 423 PPRFRRSPSYGRRSYSPRGRSPRRRSVSPRGRSYSRSPPPYRGREEVP 566
>BQ143793 weakly similar to GP|6523547|emb| hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (19%)
Length = 1100
Score = 43.9 bits (102), Expect = 7e-05
Identities = 44/166 (26%), Positives = 58/166 (34%), Gaps = 24/166 (14%)
Frame = -1
Query: 102 TGARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAG 161
TGAR ++ N+ G E + K G+ + + G G
Sbjct: 518 TGARGGGGQAKERKRGRKRRKGENAKREKGGKERGKKE--RKKGAGNGIYEGEYKGGGGG 345
Query: 162 FLNRIKKNESV------EVL----------RSGSGGGGGSSGSSKGGGGGG----TNTKE 201
R KK E E++ G GGGGG G + GGGGG KE
Sbjct: 344 GGKRKKKREGGRGKGKDEII*EKGGGMGRGNGGEGGGGGEGGGREWGGGGGDK*SLKRKE 165
Query: 202 QKK----VVNNGKGEKGKERASRYNDGGGSGSGDKRNKNIESNSKR 243
++K + GKG G E GG G+ R K E KR
Sbjct: 164 KEKDGGDKIG*GKGRGGGEGGMDVMGNGGGRRGEGRKK--EEREKR 33
Score = 41.6 bits (96), Expect = 3e-04
Identities = 31/109 (28%), Positives = 46/109 (41%), Gaps = 4/109 (3%)
Frame = -2
Query: 174 VLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKR 233
+ R + GG S K GGG G + K+ + GKG KG+E + G G
Sbjct: 706 IFREDASWGGASKTKKKEGGGRGKG-RRGKRGWDRGKGSKGQETENGRRK*GKVGKRKGE 530
Query: 234 NK----NIESNSKRNVGRPPKKAAETVASTKRGRESSASGGKDKRPKKR 278
+K E KR GR K + T+RG+ GG+ + ++R
Sbjct: 529 SKWEQGQGEGGDKRKKGRGEGKGGK--GKTQRGKRGGKKGGRRRGRRER 389
Score = 38.9 bits (89), Expect = 0.002
Identities = 33/110 (30%), Positives = 47/110 (42%), Gaps = 15/110 (13%)
Frame = -2
Query: 148 EGVSDK-KKNNGAAGFLNRIK----KNESVEVLRSGSGGGGGSSGSSKGGGGGGTN---- 198
EG +K KK G R+K + E G GGG G G GGGG N
Sbjct: 352 EGGGEKGKKKERVGGERGRMK*YRRREGGWEEEMGGRGGGEEKGGGGNGEGGGGINDL*R 173
Query: 199 TKEQKKV--VNNGKGEKGKERASRY----NDGGGSGSGDKRNKNIESNSK 242
+++KK+ + G+G+ G E + GGG G G K+ + + K
Sbjct: 172 ERKRKKMGGIR*GRGKGGGEGKGGWMLWGMGGGGGGKGGKKRRGKKGGKK 23
Score = 36.2 bits (82), Expect = 0.014
Identities = 30/111 (27%), Positives = 42/111 (37%), Gaps = 12/111 (10%)
Frame = -3
Query: 180 GGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGD-----KRN 234
GGGGG G +G G + +++ ++ E+ R N G G G +
Sbjct: 651 GGGGGRGGGGRGDGIVVREVRVKRRKMDGANEERWGRERGRVNGNRGKGRGGTSERKEEG 472
Query: 235 KNIESNSKRNVGRPPKK---AAETVASTK----RGRESSASGGKDKRPKKR 278
K E KR G+ ++ E S K RG GG K KKR
Sbjct: 471 KEKEERGKRKEGKGGERKGEEGEEEGSGKWDI*RGV*GGGRGGGKKEKKKR 319
Score = 35.0 bits (79), Expect = 0.032
Identities = 35/132 (26%), Positives = 51/132 (38%), Gaps = 1/132 (0%)
Frame = -2
Query: 146 KAEGVSDKKKNNGAAGF-LNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKK 204
K EG K G G+ + K + E R G G G SK G G ++KK
Sbjct: 661 KKEGGGRGKGRRGKRGWDRGKGSKGQETENGRRK*GKVGKRKGESKWEQGQGEGGDKRKK 482
Query: 205 VVNNGKGEKGKERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKAAETVASTKRGRE 264
GKG KGK + + GG G +R + +R +G ++G++
Sbjct: 481 GRGEGKGGKGKTQRGK---RGGKKGGRRRGRR-----EREMGYMKGSIRGGEGGGEKGKK 326
Query: 265 SSASGGKDKRPK 276
GG+ R K
Sbjct: 325 KERVGGERGRMK 290
Score = 33.9 bits (76), Expect = 0.072
Identities = 20/64 (31%), Positives = 27/64 (41%)
Frame = -1
Query: 167 KKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGG 226
K+ + + + G G GGG G G GGG G+G K +ER R +G G
Sbjct: 164 KEKDGGDKIG*GKGRGGGEGGMDVMGNGGG----------RRGEGRKKEEREKRGEEGEG 15
Query: 227 SGSG 230
G
Sbjct: 14 KEGG 3
Score = 33.9 bits (76), Expect = 0.072
Identities = 36/138 (26%), Positives = 48/138 (34%), Gaps = 27/138 (19%)
Frame = -1
Query: 171 SVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEK------GKERASRYND- 223
+ ++ + G GG G+ GGGG KE+K+ KGE GKER +
Sbjct: 569 TAQMRKGGEEKGGE*MGTGARGGGG--QAKERKRGRKRRKGENAKREKGGKERGKKERKK 396
Query: 224 -----------GGGSGSGDKRNKNIESNS---------KRNVGRPPKKAAETVASTKRGR 263
GG G G KR K E ++ G E + G
Sbjct: 395 GAGNGIYEGEYKGGGGGGGKRKKKREGGRGKGKDEII*EKGGGMGRGNGGEGGGGGEGGG 216
Query: 264 ESSASGGKDKRPKKRSKK 281
GG DK KR +K
Sbjct: 215 REWGGGGGDK*SLKRKEK 162
Score = 33.1 bits (74), Expect = 0.12
Identities = 30/106 (28%), Positives = 42/106 (39%)
Frame = -1
Query: 146 KAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKV 205
K G ++KK G N I + E + G GGGG +GG G G + +K
Sbjct: 425 KERGKKERKKGAG-----NGIYEGE----YKGGGGGGGKRKKKREGGRGKGKDEII*EKG 273
Query: 206 VNNGKGEKGKERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKK 251
G+G G +GGG G G R K ++ R K+
Sbjct: 272 GGMGRGNGG--------EGGGGGEGGGREWGGGGGDK*SLKRKEKE 159
Score = 32.0 bits (71), Expect = 0.27
Identities = 23/75 (30%), Positives = 34/75 (44%), Gaps = 7/75 (9%)
Frame = -2
Query: 148 EGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGG-------GGGGTNTK 200
+G + G L R +K + + +R G G GGG KGG GGGG
Sbjct: 226 KGGGGNGEGGGGINDL*RERKRKKMGGIR*GRGKGGGEG---KGGWMLWGMGGGGGGKGG 56
Query: 201 EQKKVVNNGKGEKGK 215
++++ GK E+GK
Sbjct: 55 KKRRGKKGGKKERGK 11
Score = 31.6 bits (70), Expect = 0.36
Identities = 36/153 (23%), Positives = 58/153 (37%), Gaps = 14/153 (9%)
Frame = -2
Query: 95 KAKEKPVTG-ARSRRSNKNLSSNASNK-KPPSNSTPRTGSSANKPAETPKLKNKAEGVSD 152
K K+K G + RR + +K + N + G + E+ + + EG
Sbjct: 670 KTKKKEGGGRGKGRRGKRGWDRGKGSKGQETENGRRK*GKVGKRKGESKWEQGQGEGGDK 491
Query: 153 KKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSS------GSSKGGGGGGTNTKEQKKVV 206
+KK G + K ++ R G GG G KG GG E+ K
Sbjct: 490 RKKGRGEG----KGGKGKTQRGKRGGKKGGRRRGRREREMGYMKGSIRGGEGGGEKGKKK 323
Query: 207 NNGKGEKGKERASRYNDG------GGSGSGDKR 233
GE+G+ + R +G GG G G+++
Sbjct: 322 ERVGGERGRMK*YRRREGGWEEEMGGRGGGEEK 224
Score = 30.0 bits (66), Expect = 1.0
Identities = 34/132 (25%), Positives = 49/132 (36%), Gaps = 26/132 (19%)
Frame = -3
Query: 165 RIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQ---------KKVVNNG---KGE 212
R+K+ + G G ++G G GGT+ +++ K+ G KGE
Sbjct: 591 RVKRRKMDGANEERWGRERGRVNGNRGKGRGGTSERKEEGKEKEERGKRKEGKGGERKGE 412
Query: 213 KGKERASRYND------GGGSGSGDKRNKNIESNSK----RNVGR----PPKKAAETVAS 258
+G+E S D GGG G G K K K N+G +K
Sbjct: 411 EGEEEGSGKWDI*RGV*GGGRGGGKKEKKKRGWEGKGEG*NNIGEGRGDGKRKWGGGGGG 232
Query: 259 TKRGRESSASGG 270
+RG E GG
Sbjct: 231 RRRGGEGMGRGG 196
>BQ151366 similar to GP|20146300|dbj hypothetical protein~similar to Oryza
sativa OSJNBa0058E19.20, partial (9%)
Length = 1137
Score = 43.9 bits (102), Expect = 7e-05
Identities = 36/140 (25%), Positives = 63/140 (44%), Gaps = 12/140 (8%)
Frame = +3
Query: 154 KKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGG---GG-----GTNTKEQKKV 205
K+N+ G K+ S +G G + GS +G G GG GT +++KK
Sbjct: 6 KENSDRKGSAREASKSRST----TGGAGRARARGSQRGAGQQDGGRRRQRGTTREKEKKG 173
Query: 206 VNNGKGEKGKERASRYNDGGGSGSGDKRN----KNIESNSKRNVGRPPKKAAETVASTKR 261
G G K++A+ + GG G G++RN E +KR +P + A +S
Sbjct: 174 A-PGPGPPAKKKAASRSGGGARGGGEQRNPRQRTKKEKKNKRKTNQPKTEGARQASSNHP 350
Query: 262 GRESSASGGKDKRPKKRSKK 281
+ A ++ RP++++ +
Sbjct: 351 PPPNQAQ--REGRPERKAHR 404
Score = 40.0 bits (92), Expect = 0.001
Identities = 46/204 (22%), Positives = 71/204 (34%), Gaps = 5/204 (2%)
Frame = +2
Query: 82 KPSYSETDEEEPPKAKEKPVTGARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETP 141
+P+ T PP P TG R+ K + +K S R G A+ P
Sbjct: 95 EPAGRGTARRRPP-----PPTGDHERKRKKRRTRPGPPRKEKSGEPKRRGGPGRGGAKKP 259
Query: 142 KLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGS-----GGGGGSSGSSKGGGGGG 196
+ NK EG +KK+ A + + + + G+ G G GS KGGG
Sbjct: 260 EATNK-EGEEKQKKDEPAQDRRGKASEQQPPPPAQPGAKRRTAGTKGPPRGSKKGGGNAD 436
Query: 197 TNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKAAETV 256
++K+ + EK R K E+N +R
Sbjct: 437 RREGQRKRRHRTARREK-----------------HPRAKTRENNPQR------------- 526
Query: 257 ASTKRGRESSASGGKDKRPKKRSK 280
KRG + S K +R K+ ++
Sbjct: 527 ---KRGAREAGSTRKKERQKRHAR 589
Score = 29.6 bits (65), Expect = 1.4
Identities = 36/194 (18%), Positives = 69/194 (35%), Gaps = 5/194 (2%)
Frame = +3
Query: 91 EEPPKAKEKPVTGARSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNKAEGV 150
++PP A R R + +S +A P+N P+ + PA + +
Sbjct: 585 QDPPPAPRGSRPQTRQNRERRRVSLSAPAHTAPTNVEPKKSRRNDAPATKRRTRVTKASR 764
Query: 151 SDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGK 210
+D + A K + +R+ ++K + K +
Sbjct: 765 ADAQNTQHTA------KVSTGDTTVRTAISHPQPKQRAAKNNRA----NERSKPYEEERR 914
Query: 211 GEK-----GKERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKAAETVASTKRGRES 265
G+K +E+A+R + G + D N I ++ R ++A E A + R
Sbjct: 915 GKKTTRRGAREKAARDSAGARADKRDTTNHRIRYQNQPTK*RAEREAEEKSAE*ENERRP 1094
Query: 266 SASGGKDKRPKKRS 279
+A R +KR+
Sbjct: 1095AARATISTREEKRT 1136
>BQ141541 similar to GP|334068|gb|A ORF2 {Pseudorabies virus}, partial (2%)
Length = 1320
Score = 43.5 bits (101), Expect = 9e-05
Identities = 39/134 (29%), Positives = 59/134 (43%), Gaps = 11/134 (8%)
Frame = +3
Query: 105 RSRRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDK-----KKNNGA 159
+ R++ N++ N +N++ + + + N A K + +AEG + KK G
Sbjct: 402 KKRKN*YNITGNGTNEQ--TRTEANENEAENGRATRRK*RGRAEGRRREGDP*GKKGGGR 575
Query: 160 AGFLNRIKKNESVEVLRSGSGGGGGSSGS------SKGGGGGGTNTKEQKKVVNNGKGEK 213
AG L E R G GGGG G+ S+GGGG +KK G G+
Sbjct: 576 AGGL---------EGRRGGG*GGGGRGGAGAKRRRSRGGGGRIATAGSRKK--GGGVGDA 722
Query: 214 GKERASRYNDGGGS 227
G+ER R + G S
Sbjct: 723 GRERHERRGENGRS 764
Score = 30.8 bits (68), Expect = 0.61
Identities = 41/181 (22%), Positives = 73/181 (39%), Gaps = 12/181 (6%)
Frame = +1
Query: 103 GARSRRSNKNLSSNAS-NKKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAG 161
G R+R++ K +N + + P++ R + + + + +N+ G ++ +G G
Sbjct: 379 GRRTRKTIKKEKTNTT*QEMAPTSKRERKRTKTRQKMDERREENEGGG----QRGDGGKG 546
Query: 162 FLNRIKKNESVEVLRSGSGGGGGSSGSSKGGGGG-----GTNTKEQKK-VVNNGKGE--- 212
+++ KK GG +GGGGG G +E K+ V +GE
Sbjct: 547 -IHKEKKE-------------GGGPAVWRGGGGGGRGEVGEGARELKEGGVGGEEGE*LP 684
Query: 213 --KGKERASRYNDGGGSGSGDKRNKNIESNSKRNVGRPPKKAAETVASTKRGRESSASGG 270
G++ G SG +R ++ R R ++ E AS R SA GG
Sbjct: 685 RAAGRKAEGLVTRGERDTSGGERTED-RKYEHRIAERERRREGEKEASNTRREARSARGG 861
Query: 271 K 271
+
Sbjct: 862 R 864
>TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 1054
Score = 43.1 bits (100), Expect = 1e-04
Identities = 23/57 (40%), Positives = 26/57 (45%)
Frame = +1
Query: 177 SGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKR 233
SG GGGGG G GGGGGG G +G Y GGG G G++R
Sbjct: 598 SGGGGGGGRGGGGYGGGGGG-----------YGGERRGYGGGGGYGGGGGGGYGERR 735
Score = 32.0 bits (71), Expect = 0.27
Identities = 13/21 (61%), Positives = 14/21 (65%)
Frame = +1
Query: 176 RSGSGGGGGSSGSSKGGGGGG 196
R G GGG G G S+GGG GG
Sbjct: 757 RGGGGGGYGGGGYSRGGGDGG 819
Score = 31.6 bits (70), Expect = 0.36
Identities = 19/49 (38%), Positives = 22/49 (44%)
Frame = +1
Query: 180 GGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSG 228
GGGGG G +GGGGG + G G G Y+ GGG G
Sbjct: 703 GGGGGGYGERRGGGGGYSR--------GGGGGGYG---GGGYSRGGGDG 816
>TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 754
Score = 43.1 bits (100), Expect = 1e-04
Identities = 23/57 (40%), Positives = 26/57 (45%)
Frame = +2
Query: 177 SGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKR 233
SG GGGGG G GGGGGG G +G Y GGG G G++R
Sbjct: 344 SGGGGGGGRGGGGYGGGGGG-----------YGGERRGYGGGGGYGGGGGGGYGERR 481
Score = 32.0 bits (71), Expect = 0.27
Identities = 13/21 (61%), Positives = 14/21 (65%)
Frame = +2
Query: 176 RSGSGGGGGSSGSSKGGGGGG 196
R G GGG G G S+GGG GG
Sbjct: 503 RGGGGGGYGGGGYSRGGGDGG 565
Score = 31.6 bits (70), Expect = 0.36
Identities = 19/49 (38%), Positives = 22/49 (44%)
Frame = +2
Query: 180 GGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSG 228
GGGGG G +GGGGG + G G G Y+ GGG G
Sbjct: 449 GGGGGGYGERRGGGGGYSR--------GGGGGGYG---GGGYSRGGGDG 562
>TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 912
Score = 43.1 bits (100), Expect = 1e-04
Identities = 23/57 (40%), Positives = 26/57 (45%)
Frame = +1
Query: 177 SGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSGSGDKR 233
SG GGGGG G GGGGGG G +G Y GGG G G++R
Sbjct: 502 SGGGGGGGRGGGGYGGGGGG-----------YGGERRGYGGGGGYGGGGGGGYGERR 639
Score = 32.0 bits (71), Expect = 0.27
Identities = 13/21 (61%), Positives = 14/21 (65%)
Frame = +1
Query: 176 RSGSGGGGGSSGSSKGGGGGG 196
R G GGG G G S+GGG GG
Sbjct: 661 RGGGGGGYGGGGYSRGGGDGG 723
Score = 31.6 bits (70), Expect = 0.36
Identities = 19/49 (38%), Positives = 22/49 (44%)
Frame = +1
Query: 180 GGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYNDGGGSG 228
GGGGG G +GGGGG + G G G Y+ GGG G
Sbjct: 607 GGGGGGYGERRGGGGGYSR--------GGGGGGYG---GGGYSRGGGDG 720
>TC79086 similar to GP|23499033|emb|CAD51113. ubiquitin-protein ligase 1
putative {Plasmodium falciparum 3D7}, partial (0%)
Length = 1418
Score = 42.7 bits (99), Expect = 2e-04
Identities = 30/130 (23%), Positives = 53/130 (40%), Gaps = 10/130 (7%)
Frame = +2
Query: 57 SHKGDHKPIINDKKERPSSDVKPTLKPSYS----------ETDEEEPPKAKEKPVTGARS 106
S +G + + ++E P + P P + +TDE+EPP K KP+
Sbjct: 158 SSEGGEEEEESSEEEAPKTTPPPKTNPKNNSVEDEDEEETDTDEDEPP-PKTKPIEQTLK 334
Query: 107 RRSNKNLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRI 166
+ +++ S K P+ + S+ K E K + E D NN A F
Sbjct: 335 SGTKPSIAPARSGTKRPAENNDTKQSNKKKTTEEKNKKKEKEPEEDDNSNNKKASFQRVF 514
Query: 167 KKNESVEVLR 176
+++ V +L+
Sbjct: 515 TEDDEVAILQ 544
>TC91646 weakly similar to GP|6523547|emb|CAB62280.1 hydroxyproline-rich
glycoprotein DZ-HRGP {Volvox carteri f. nagariensis},
partial (15%)
Length = 984
Score = 41.6 bits (96), Expect = 3e-04
Identities = 33/101 (32%), Positives = 41/101 (39%), Gaps = 3/101 (2%)
Frame = -2
Query: 134 ANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNESVEVLRSGSGGGGGSSGSSKG-G 192
A + + L G D+ A G KK E+++ R G GGGGSS S G G
Sbjct: 746 AGRQSSNAALDGHGVGGEDQVVGGEAGGRTEAAKKREAIDEGRVGGNGGGGSSRLSAGRG 567
Query: 193 GGGGTNTKEQKKVVNNGKGEKGKERAS--RYNDGGGSGSGD 231
GG T V G+G G R + GG G GD
Sbjct: 566 GGRDTGGVAAGGDVGEGEGLIGGRGVGVVRDSSGGFLGGGD 444
>BQ150123 similar to GP|21109992|gb| TonB-dependent receptor {Xanthomonas
axonopodis pv. citri str. 306}, partial (2%)
Length = 1131
Score = 40.4 bits (93), Expect = 8e-04
Identities = 34/127 (26%), Positives = 48/127 (37%), Gaps = 24/127 (18%)
Frame = +2
Query: 178 GSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYND---------GGGSG 228
G GGGGG+ + K G G E K G G++G++ D G GSG
Sbjct: 8 GRGGGGGTEKTEKRNGREGRKRTEGK----GGGGKEGQKNKRERKDTIQKAPRERGAGSG 175
Query: 229 SG----DKRNKNIESNSKRNVGRPPKKAAETVASTKRGRESSAS-----------GGKDK 273
K K E R G PP+ + A + E S S GG+++
Sbjct: 176 EAKRMEQKSTKKREQPGPRTHGTPPRPDPRSAAPQRNSAEKSRSSSPHQQQTPTQGGRNR 355
Query: 274 RPKKRSK 280
+ R+K
Sbjct: 356 ATRARAK 376
Score = 30.0 bits (66), Expect = 1.0
Identities = 25/101 (24%), Positives = 40/101 (38%), Gaps = 11/101 (10%)
Frame = +1
Query: 177 SGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRYND-----GGGSGSGD 231
+G G G + ++ G K ++K G K ++RA RY+ GG G
Sbjct: 1 TGGERGRGRNRENRKAKRQGRKKKNRRKGGRGEGGTKEQKRAERYDSKGPEGAGGGKRGG 180
Query: 232 KRNKNIESNSKRNVG------RPPKKAAETVASTKRGRESS 266
K N + R G PP + + A+ K+ R+ S
Sbjct: 181 KTNGTKKYKKTRTAGTANARNAPPTRPPQRRATKKQRRKVS 303
>CA923192 similar to GP|20196906|gb| expressed protein {Arabidopsis
thaliana}, partial (16%)
Length = 762
Score = 40.4 bits (93), Expect = 8e-04
Identities = 35/110 (31%), Positives = 44/110 (39%)
Frame = -3
Query: 112 NLSSNASNKKPPSNSTPRTGSSANKPAETPKLKNKAEGVSDKKKNNGAAGFLNRIKKNES 171
NLS + K S ST A +PA T N KK G+ G N++
Sbjct: 607 NLSWKEATK---SASTTAPAVGAKRPATTGNGSNNIA----LKKGRGSGGMQNQL----- 464
Query: 172 VEVLRSGSGGGGGSSGSSKGGGGGGTNTKEQKKVVNNGKGEKGKERASRY 221
V G GGGG G GGG GG + Q + G+G G+ R Y
Sbjct: 463 VNRALGGLSGGGGGRGRVSGGGRGGARGRGQGQGWGRGRG-SGRGRGRGY 317
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.299 0.122 0.330
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,311,785
Number of Sequences: 36976
Number of extensions: 116594
Number of successful extensions: 3862
Number of sequences better than 10.0: 366
Number of HSP's better than 10.0 without gapping: 1655
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2483
length of query: 281
length of database: 9,014,727
effective HSP length: 95
effective length of query: 186
effective length of database: 5,502,007
effective search space: 1023373302
effective search space used: 1023373302
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 17 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146756.6


