

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146756.3 - phase: 0
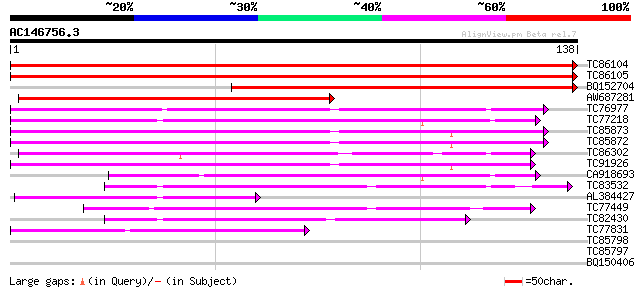
(138 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86104 homologue to SP|O48551|PSA6_SOYBN Proteasome subunit alp... 279 3e-76
TC86105 homologue to SP|O48551|PSA6_SOYBN Proteasome subunit alp... 264 7e-72
BQ152704 homologue to SP|O48551|PSA6_ Proteasome subunit alpha t... 136 3e-33
AW687281 weakly similar to SP|Q9LSU3|PSA6 Proteasome subunit alp... 102 6e-23
TC76977 homologue to SP|O23708|PSA2_ARATH Proteasome subunit alp... 97 3e-21
TC77218 similar to GP|22947842|gb|AAN07899.1 20S proteasome alph... 77 2e-15
TC85873 homologue to SP|P52427|PSA4_SPIOL Proteasome subunit alp... 72 5e-14
TC85872 homologue to SP|P52427|PSA4_SPIOL Proteasome subunit alp... 69 6e-13
TC86302 homologue to SP|Q9SXU1|PSA7_CICAR Proteasome subunit alp... 67 3e-12
TC91926 similar to SP|P52427|PSA4_SPIOL Proteasome subunit alpha... 66 5e-12
CA918693 similar to SP|P34066|PS11 Proteasome subunit alpha type... 62 1e-10
TC83532 similar to PIR|T38639|T38639 proteasome component PUP2 h... 55 1e-08
AL384427 similar to GP|18376026|em probable 20S proteasome subun... 52 8e-08
TC77449 homologue to SP|Q9M4T8|PSA5_SOYBN Proteasome subunit alp... 52 8e-08
TC82430 weakly similar to SP|Q9LSU1|PSA5_ORYSA Proteasome subuni... 52 1e-07
TC77831 homologue to GP|14594925|emb|CAC43323. putative alpha7 p... 41 2e-04
TC85798 similar to PIR|T51979|T51979 multicatalytic endopeptidas... 32 0.11
TC85797 similar to PIR|T51979|T51979 multicatalytic endopeptidas... 32 0.11
BQ150406 25 9.9
>TC86104 homologue to SP|O48551|PSA6_SOYBN Proteasome subunit alpha type 6
(EC 3.4.25.1) (20S proteasome alpha subunit A), complete
Length = 1113
Score = 279 bits (713), Expect = 3e-76
Identities = 138/138 (100%), Positives = 138/138 (100%)
Frame = +3
Query: 1 MPVDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSA 60
MPVDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSA
Sbjct: 474 MPVDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSA 653
Query: 61 GSKDQEAINFLEKKMKNDPSFTYDETVQTAIAALQSVLQEDFKATEIEVGVVQKDKPEFR 120
GSKDQEAINFLEKKMKNDPSFTYDETVQTAIAALQSVLQEDFKATEIEVGVVQKDKPEFR
Sbjct: 654 GSKDQEAINFLEKKMKNDPSFTYDETVQTAIAALQSVLQEDFKATEIEVGVVQKDKPEFR 833
Query: 121 VLSTEEIDEHLTAISERD 138
VLSTEEIDEHLTAISERD
Sbjct: 834 VLSTEEIDEHLTAISERD 887
>TC86105 homologue to SP|O48551|PSA6_SOYBN Proteasome subunit alpha type 6
(EC 3.4.25.1) (20S proteasome alpha subunit A), complete
Length = 1047
Score = 264 bits (675), Expect = 7e-72
Identities = 130/138 (94%), Positives = 134/138 (96%)
Frame = +2
Query: 1 MPVDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSA 60
MPVDVL+KW+ADKSQVYTQHAYMRPLGVVAMVLGIDDE GPQLYKCDPAGHYFGHKATSA
Sbjct: 455 MPVDVLSKWVADKSQVYTQHAYMRPLGVVAMVLGIDDELGPQLYKCDPAGHYFGHKATSA 634
Query: 61 GSKDQEAINFLEKKMKNDPSFTYDETVQTAIAALQSVLQEDFKATEIEVGVVQKDKPEFR 120
G KDQEAINFLEKKMKNDPSFTY ETVQTAI+ALQSVLQEDFKATEIEVGVVQKD PEFR
Sbjct: 635 GLKDQEAINFLEKKMKNDPSFTYAETVQTAISALQSVLQEDFKATEIEVGVVQKDNPEFR 814
Query: 121 VLSTEEIDEHLTAISERD 138
VLST+EIDEHLTAISERD
Sbjct: 815 VLSTDEIDEHLTAISERD 868
>BQ152704 homologue to SP|O48551|PSA6_ Proteasome subunit alpha type 6 (EC
3.4.25.1) (20S proteasome alpha subunit A), partial
(28%)
Length = 427
Score = 136 bits (342), Expect = 3e-33
Identities = 68/84 (80%), Positives = 75/84 (88%)
Frame = +2
Query: 55 HKATSAGSKDQEAINFLEKKMKNDPSFTYDETVQTAIAALQSVLQEDFKATEIEVGVVQK 114
H+A+ + + +NFLEKKMKNDPSFTY ETVQTAI+ALQSVLQEDFKATEIEVGVVQK
Sbjct: 2 HEASCPSCRIRHEVNFLEKKMKNDPSFTYAETVQTAISALQSVLQEDFKATEIEVGVVQK 181
Query: 115 DKPEFRVLSTEEIDEHLTAISERD 138
D PEFRVLST+EIDEHLTAISERD
Sbjct: 182 DNPEFRVLSTDEIDEHLTAISERD 253
>AW687281 weakly similar to SP|Q9LSU3|PSA6 Proteasome subunit alpha type 6
(EC 3.4.25.1) (20S proteasome alpha subunit A), partial
(65%)
Length = 612
Score = 102 bits (253), Expect = 6e-23
Identities = 49/77 (63%), Positives = 59/77 (75%)
Frame = +3
Query: 3 VDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSAGS 62
V +A A+ +QVYTQHA+MR LGVV + GIDDE GPQLY+ DPAGHY G KA +AG+
Sbjct: 300 VPYMANRTANVAQVYTQHAFMRALGVVTIFCGIDDEKGPQLYRSDPAGHYVGFKACAAGA 479
Query: 63 KDQEAINFLEKKMKNDP 79
K+QEA NFLEKK+K P
Sbjct: 480 KEQEANNFLEKKIKATP 530
>TC76977 homologue to SP|O23708|PSA2_ARATH Proteasome subunit alpha type 2
(EC 3.4.25.1) (20S proteasome alpha subunit B).
[Mouse-ear cress], complete
Length = 1150
Score = 96.7 bits (239), Expect = 3e-21
Identities = 55/131 (41%), Positives = 77/131 (57%)
Frame = +3
Query: 1 MPVDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSA 60
+PV L + +A Q +TQ +RP GV +V G DD NGPQLY+ DP+G YF KA++
Sbjct: 417 IPVTQLVREVAAVMQEFTQSGGVRPFGVSLLVAGFDD-NGPQLYQVDPSGSYFSWKASAM 593
Query: 61 GSKDQEAINFLEKKMKNDPSFTYDETVQTAIAALQSVLQEDFKATEIEVGVVQKDKPEFR 120
G A FLEK+ +D D+ V TAI L+ + A IE+G++ DK +FR
Sbjct: 594 GKNVSNAKTFLEKRYTDD--MELDDAVHTAILTLKEGFEGQISAKNIEIGIIGADK-KFR 764
Query: 121 VLSTEEIDEHL 131
VL+ EID++L
Sbjct: 765 VLTPAEIDDYL 797
>TC77218 similar to GP|22947842|gb|AAN07899.1 20S proteasome alpha 6 subunit
{Nicotiana benthamiana}, partial (91%)
Length = 1236
Score = 77.0 bits (188), Expect = 2e-15
Identities = 45/130 (34%), Positives = 77/130 (58%), Gaps = 1/130 (0%)
Frame = +2
Query: 1 MPVDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSA 60
+PV L +ADK+QV TQ ++ RP GV +V G+D E+G LY P+G+YF ++A +
Sbjct: 377 LPVGRLVVQLADKAQVCTQRSWKRPYGVGLLVAGLD-ESGAHLYYNCPSGNYFEYQAFAI 553
Query: 61 GSKDQEAINFLEKKMKNDPSFTYDETVQTAIAALQSVLQ-EDFKATEIEVGVVQKDKPEF 119
GS+ Q A +LE+K N + + ++ ++ A+ A + LQ E +++ + VV +P F
Sbjct: 554 GSRSQAAKTYLERKFNNFTASSREDLIKDALIATRESLQGEKLRSSVCTIAVVGVGEP-F 730
Query: 120 RVLSTEEIDE 129
+L E + +
Sbjct: 731 HILDQETVQQ 760
>TC85873 homologue to SP|P52427|PSA4_SPIOL Proteasome subunit alpha type 4
(EC 3.4.25.1) (20S proteasome alpha subunit C), partial
(98%)
Length = 1042
Score = 72.4 bits (176), Expect = 5e-14
Identities = 46/135 (34%), Positives = 71/135 (52%), Gaps = 4/135 (2%)
Frame = +2
Query: 1 MPVDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSA 60
MPV+ L + + D Q YTQ +RP GV + G D G QLY DP+G+Y G KA++
Sbjct: 383 MPVEQLVQSLCDTKQGYTQFGGLRPFGVSFLFAGWDKNFGFQLYMSDPSGNYGGWKASAI 562
Query: 61 GSKDQEAINFLEKKMKNDPSFTYDETVQTAIAALQSVLQEDFKATE----IEVGVVQKDK 116
G+ +Q A + L++ K+D T +E VQ A+ L + +E EV + K
Sbjct: 563 GANNQAAQSILKQDYKDD--ITREEAVQLALKVLSKTMDSTSLTSEKLELAEVFLSPSGK 736
Query: 117 PEFRVLSTEEIDEHL 131
+++V S E + + L
Sbjct: 737 VKYQVCSPENLTKLL 781
>TC85872 homologue to SP|P52427|PSA4_SPIOL Proteasome subunit alpha type 4
(EC 3.4.25.1) (20S proteasome alpha subunit C), complete
Length = 1139
Score = 68.9 bits (167), Expect = 6e-13
Identities = 44/135 (32%), Positives = 69/135 (50%), Gaps = 4/135 (2%)
Frame = +2
Query: 1 MPVDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSA 60
MPV+ L + + D Q YTQ +RP GV + G D G QLY DP+G+Y G KA +
Sbjct: 440 MPVEQLVQSLCDTKQGYTQFGGLRPFGVSFLFAGWDKNFGFQLYMSDPSGNYGGWKAGAI 619
Query: 61 GSKDQEAINFLEKKMKNDPSFTYDETVQTAIAALQSVLQEDFKATE----IEVGVVQKDK 116
G+ +Q A + L++ K+D T +E V A+ L + ++ EV + K
Sbjct: 620 GANNQAAQSILKQDYKDD--ITREEAVNLALKVLSKTMDSTSLTSDKLELAEVFLAPSGK 793
Query: 117 PEFRVLSTEEIDEHL 131
+++V S E + + L
Sbjct: 794 VKYQVCSPENLTKLL 838
>TC86302 homologue to SP|Q9SXU1|PSA7_CICAR Proteasome subunit alpha type 7
(EC 3.4.25.1) (20S proteasome alpha subunit D), complete
Length = 1326
Score = 66.6 bits (161), Expect = 3e-12
Identities = 46/127 (36%), Positives = 63/127 (49%), Gaps = 1/127 (0%)
Frame = +2
Query: 3 VDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENG-PQLYKCDPAGHYFGHKATSAG 61
V+ + + IA Q YTQ +RP G+ +++G D G P LY+ DP+G + KA + G
Sbjct: 635 VEYITRHIAGLQQKYTQSGGVRPFGLSTLIVGFDPYTGSPSLYQTDPSGTFSAWKANATG 814
Query: 62 SKDQEAINFLEKKMKNDPSFTYDETVQTAIAALQSVLQEDFKATEIEVGVVQKDKPEFRV 121
FLEK K ETV+ AI AL V++ K IEV V+ K+ R
Sbjct: 815 RNSNSIREFLEKNFKETSG---QETVKLAIRALLEVVESGGK--NIEVAVMTKEH-GLRQ 976
Query: 122 LSTEEID 128
L EID
Sbjct: 977 LEEAEID 997
>TC91926 similar to SP|P52427|PSA4_SPIOL Proteasome subunit alpha type 4 (EC
3.4.25.1) (20S proteasome alpha subunit C), partial
(78%)
Length = 728
Score = 65.9 bits (159), Expect = 5e-12
Identities = 41/132 (31%), Positives = 66/132 (49%), Gaps = 4/132 (3%)
Frame = +1
Query: 1 MPVDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSA 60
+PV+ L + + D Q YTQ+ +RP GV + G D G QLY DP+G+Y G KAT
Sbjct: 265 IPVEQLVQRLCDLKQGYTQYGGLRPFGVSFIYAGHDSHYGFQLYHSDPSGNYGGWKATCI 444
Query: 61 GSKDQEAINFLEKKMKNDPSFTYDETVQTAIAALQSVLQEDFKATE----IEVGVVQKDK 116
G+ + A + L++ K+D T E + A L + ++E + + ++K
Sbjct: 445 GANNASAQSLLKQDYKDD--ITLKEAKELAAKVLSKTMDSTTLSSEKLEFATISLNDENK 618
Query: 117 PEFRVLSTEEID 128
+ + EEID
Sbjct: 619 VVYHLYKPEEID 654
>CA918693 similar to SP|P34066|PS11 Proteasome subunit alpha type 1-1 (EC
3.4.25.1) (20S proteasome alpha subunit F1), partial
(43%)
Length = 680
Score = 61.6 bits (148), Expect = 1e-10
Identities = 33/106 (31%), Positives = 61/106 (57%), Gaps = 1/106 (0%)
Frame = -2
Query: 25 PLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSAGSKDQEAINFLEKKMKNDPSFTYD 84
P GVV +V G+D+ Y C P+G+YF ++A + S+ Q A +LE+K N + + +
Sbjct: 679 PYGVVLLVAGLDESEAHLYYNC-PSGNYFEYQAVAIDSRSQAAKTYLERKFNNFTASSRE 503
Query: 85 ETVQTAIAALQSVLQ-EDFKATEIEVGVVQKDKPEFRVLSTEEIDE 129
+ ++ A+ A + LQ E+ +++ + VV D+P F +L E + +
Sbjct: 502 DLIKDALIATRESLQGEELRSSVCTIAVVGVDEP-FHILDQETVQQ 368
>TC83532 similar to PIR|T38639|T38639 proteasome component PUP2 homolog -
fission yeast (Schizosaccharomyces pombe), partial (97%)
Length = 901
Score = 54.7 bits (130), Expect = 1e-08
Identities = 34/114 (29%), Positives = 62/114 (53%)
Frame = +3
Query: 24 RPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSAGSKDQEAINFLEKKMKNDPSFTY 83
RP GV ++ G D E+GPQL+ +P+G ++ + A + GS + A L+ + + T
Sbjct: 480 RPFGVALLIAGYD-EDGPQLFHAEPSGTFYRYDAKAIGSGSEGAQAELQNEYHKSLTITD 656
Query: 84 DETVQTAIAALQSVLQEDFKATEIEVGVVQKDKPEFRVLSTEEIDEHLTAISER 137
ET+ + L+ V++E + +++ V K+K FR+ + DE + A+ ER
Sbjct: 657 AETL--VLKTLKQVMEEKLDSKNVQLASVTKEK-GFRIYT----DEEMAAVVER 797
>AL384427 similar to GP|18376026|em probable 20S proteasome subunit Y7
{Neurospora crassa}, partial (59%)
Length = 462
Score = 52.0 bits (123), Expect = 8e-08
Identities = 25/60 (41%), Positives = 36/60 (59%)
Frame = +2
Query: 2 PVDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSAG 61
P +L K +A Q +TQ +RP GV ++ G D E GP LY+ DP+G Y+ KA++ G
Sbjct: 281 PTGILVKEVASVMQEFTQSGGVRPFGVSLLIAGYD-ETGPALYQVDPSGSYWAWKASAIG 457
>TC77449 homologue to SP|Q9M4T8|PSA5_SOYBN Proteasome subunit alpha type 5
(EC 3.4.25.1) (20S proteasome alpha subunit E), complete
Length = 1091
Score = 52.0 bits (123), Expect = 8e-08
Identities = 30/110 (27%), Positives = 58/110 (52%)
Frame = +2
Query: 19 QHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSAGSKDQEAINFLEKKMKND 78
+ + RP GV ++ G DENGP LY DP+G ++ A + GS + A + L+++ D
Sbjct: 479 EESMSRPFGVSLLIAG-HDENGPSLYYTDPSGTFWQCNAKAIGSGSEGADSSLQEQFNKD 655
Query: 79 PSFTYDETVQTAIAALQSVLQEDFKATEIEVGVVQKDKPEFRVLSTEEID 128
+ ET+ A++ L+ V++E +++ V P + + + E++
Sbjct: 656 LTLQEAETI--ALSILKQVMEEKVTPNNVDIARV---APAYHLYTPSEVE 790
>TC82430 weakly similar to SP|Q9LSU1|PSA5_ORYSA Proteasome subunit alpha
type 5 (EC 3.4.25.1) (20S proteasome alpha subunit E),
partial (77%)
Length = 561
Score = 51.6 bits (122), Expect = 1e-07
Identities = 29/89 (32%), Positives = 48/89 (53%)
Frame = +1
Query: 24 RPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSAGSKDQEAINFLEKKMKNDPSFTY 83
RP GV +V GID E GPQL+ DP+G + + A + GS + A L+++ S T
Sbjct: 304 RPFGVALLVAGID-EKGPQLFHTDPSGTFMKYNAKAIGSGSEGAQTELQEEYHK--SITL 474
Query: 84 DETVQTAIAALQSVLQEDFKATEIEVGVV 112
+E ++ L+ V++E T +++ V
Sbjct: 475 EEAETLSLKVLKQVMEEKLNNTNVQLASV 561
>TC77831 homologue to GP|14594925|emb|CAC43323. putative alpha7 proteasome
subunit {Nicotiana tabacum}, complete
Length = 1109
Score = 40.8 bits (94), Expect = 2e-04
Identities = 24/73 (32%), Positives = 39/73 (52%)
Frame = +1
Query: 1 MPVDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSA 60
+PV LA +A + T + ++RP G ++LG D +GPQLY +P+G + + +
Sbjct: 397 IPVKELADRVASYVHLCTLYWWLRPFGC-GVILGGYDRDGPQLYMVEPSGVSYRYFGAAI 573
Query: 61 GSKDQEAINFLEK 73
G Q A +EK
Sbjct: 574 GKGRQAAKTEIEK 612
>TC85798 similar to PIR|T51979|T51979 multicatalytic endopeptidase complex
(EC 3.4.99.46) chain PBB2 [imported] - Arabidopsis
thaliana, partial (94%)
Length = 1161
Score = 31.6 bits (70), Expect = 0.11
Identities = 25/91 (27%), Positives = 42/91 (45%), Gaps = 1/91 (1%)
Frame = +1
Query: 30 AMVLGIDDENGPQLYKCDPAGHYFGHKATSAGSKDQEAINFLEKKMKNDPSFTYDETVQT 89
A+VLG D GP L+ P G + GS A++ E K K S + DE V+
Sbjct: 514 ALVLGGVDITGPHLHTIYPHGSTDTLPFATMGSGSLAAMSVFESKYKE--SLSRDEGVKL 687
Query: 90 AIAALQSVLQEDF-KATEIEVGVVQKDKPEF 119
+ ++ + + D + ++V V+ K E+
Sbjct: 688 VVESICAGIFNDLGSGSNVDVCVITKGHKEY 780
>TC85797 similar to PIR|T51979|T51979 multicatalytic endopeptidase complex
(EC 3.4.99.46) chain PBB2 [imported] - Arabidopsis
thaliana, partial (94%)
Length = 1542
Score = 31.6 bits (70), Expect = 0.11
Identities = 25/91 (27%), Positives = 42/91 (45%), Gaps = 1/91 (1%)
Frame = +1
Query: 30 AMVLGIDDENGPQLYKCDPAGHYFGHKATSAGSKDQEAINFLEKKMKNDPSFTYDETVQT 89
A+VLG D GP L+ P G + GS A++ E K K S + DE V+
Sbjct: 862 ALVLGGVDITGPHLHTIYPHGSTDTLPFATMGSGSLAAMSVFESKYKE--SLSRDEGVKL 1035
Query: 90 AIAALQSVLQEDF-KATEIEVGVVQKDKPEF 119
+ ++ + + D + ++V V+ K E+
Sbjct: 1036VVESICAGIFNDLGSGSNVDVCVITKGHKEY 1128
>BQ150406
Length = 468
Score = 25.0 bits (53), Expect = 9.9
Identities = 12/30 (40%), Positives = 19/30 (63%)
Frame = -3
Query: 108 EVGVVQKDKPEFRVLSTEEIDEHLTAISER 137
E+GV++K + FR+ +ID+ T IS R
Sbjct: 280 EMGVIEKFERRFRICEVYDIDDTNT*ISIR 191
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.314 0.132 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,269,747
Number of Sequences: 36976
Number of extensions: 34049
Number of successful extensions: 106
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 97
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 98
length of query: 138
length of database: 9,014,727
effective HSP length: 86
effective length of query: 52
effective length of database: 5,834,791
effective search space: 303409132
effective search space used: 303409132
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 53 (25.0 bits)
Medicago: description of AC146756.3


