

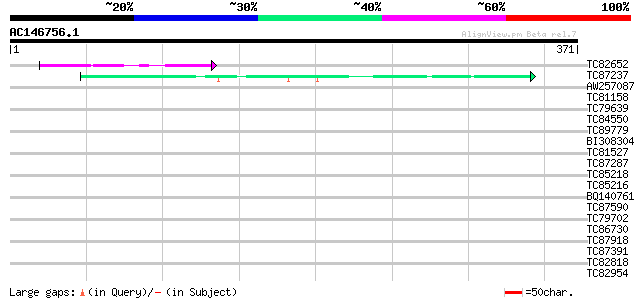
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146756.1 - phase: 0
(371 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC82652 similar to PIR|B96638|B96638 hypothetical protein F11P17... 68 6e-12
TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-inf... 42 3e-04
AW257087 homologue to GP|6683126|dbj KIAA0339 protein {Homo sapi... 36 0.027
TC81158 similar to GP|9280664|gb|AAF86533.1| F21B7.15 {Arabidops... 34 0.079
TC79639 similar to GP|10728064|gb|AAF50455.2 CG7060 gene product... 32 0.30
TC84550 weakly similar to PIR|T46095|T46095 hypothetical protein... 32 0.30
TC89779 GP|12835923|dbj|BAB23419. data source:MGD source key:MG... 32 0.39
BI308304 similar to GP|18642468|em transcriptional adaptor {Zea ... 32 0.51
TC81527 similar to GP|9954116|gb|AAG08961.1| tuber-specific and ... 31 0.67
TC87287 similar to GP|7546725|gb|AAF63657.1| MFP1 attachment fac... 31 0.87
TC85218 similar to SP|P05692|LEGJ_PEA Legumin J precursor. [Gard... 31 0.87
TC85216 similar to PIR|S26688|S26688 legumin K - garden pea, par... 31 0.87
BQ140761 weakly similar to GP|18076245|emb phosphophoryn {Rattus... 31 0.87
TC87590 weakly similar to PIR|G96522|G96522 F11A17.16 [imported]... 30 1.1
TC79702 MtN14 30 1.1
TC86730 similar to GP|13811646|gb|AAK40224.1 syntaxin of plants ... 30 1.5
TC87918 weakly similar to GP|15810397|gb|AAL07086.1 unknown prot... 30 1.5
TC87391 homologue to GP|22832155|gb|AAF48195.2 CG4013-PA {Drosop... 30 1.9
TC82818 similar to GP|20259203|gb|AAM14317.1 unknown protein {Ar... 29 2.5
TC82954 weakly similar to GP|8809591|dbj|BAA97142.1 emb|CAB82814... 29 2.5
>TC82652 similar to PIR|B96638|B96638 hypothetical protein F11P17.6
[imported] - Arabidopsis thaliana, partial (6%)
Length = 735
Score = 67.8 bits (164), Expect = 6e-12
Identities = 41/116 (35%), Positives = 65/116 (55%)
Frame = +1
Query: 20 GWGTWEELLLASAVDRHGFKDWDTIAMEVQSRTNRTSLLATAHHCEQKFHDLNRRFKDDV 79
GWGTWEELLL AV R+G +DW+ +A E++ R + + T C+ KF DL +R+
Sbjct: 358 GWGTWEELLLGGAVFRYGTRDWNVVAGELRERID-CPIPFTPEVCKAKFEDLQQRY---- 522
Query: 80 PPPQQNGDVSAVTAEDSDHVPWLDKLRKQRVAELRRDVQLSDVSILSLQLQVKKLE 135
+G + ++LRK+RV EL++ ++ S SI SL+ +++ LE
Sbjct: 523 -----SGSTDFL----------YEELRKRRVEELKKAIERSGDSIGSLKSKIEDLE 645
>TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-infected
erythrocyte surface antigen {Plasmodium falciparum},
partial (2%)
Length = 2007
Score = 42.4 bits (98), Expect = 3e-04
Identities = 68/346 (19%), Positives = 121/346 (34%), Gaps = 48/346 (13%)
Frame = +1
Query: 47 EVQSRTNRTSLLATAHHCEQKFHDLNRRFKDDVPPPQQNGDVSAVTAEDSDHVPWLDKLR 106
E +S T E+K H N KD+ Q+N ++ ++ D+ +
Sbjct: 55 EEKSHLENEESKETGESSEEKSHLENEENKDEEKSKQENEEIKDGEKIQQENEENKDEEK 234
Query: 107 KQRVAELRRDVQLSDVSILSLQLQVKKLE----------DEKAK-ENEEKVETEPDLAVS 155
Q+ E +D + S + ++KK E +EK+K ENEE ET S
Sbjct: 235 SQQENEENKDEEKSQQ-----ENELKKNEGGEKETGEITEEKSKQENEETSETN-----S 384
Query: 156 GEGRLPENEKTGGDIDEPVPAIRRLD--ESTTNTDKLLPAAGEESDR------------- 200
+ E+ + G D E V D + T T+ EE D+
Sbjct: 385 KDKENEESNQNGSDAKEQVGENHEQDSKQGTEETNGTEGGEKEEHDKIKEDTSSDNQVQD 564
Query: 201 ----------------------DNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSV 238
DN+S SN T +FD + E
Sbjct: 565 GEKNNEAREENYSGDNASSAVVDNKSQESSNKTEEQFDKKEKNE---------------F 699
Query: 239 QVKSGLNEPDQTGRKRKSVEEESNNGSYDNNEAKAVTCESVPPSEERKVEDDSVTRSEGG 298
+++S N + T ++ + S + ++A+ E+ P D+ E
Sbjct: 700 ELESQKNSNETTESTDSTITQNSQGNESEKDQAQT---ENDTPKGSASESDEQKQEQEQN 870
Query: 299 GTRESSEVQSSSSLSKTRRTQRRRKEVSGEDAKMENEDVAVVKSEP 344
T + +VQ++ + S+ ++ + EDA + ED + + + P
Sbjct: 871 NTTK-DDVQTTDTSSQNGNDTTEKQNETSEDANSKKEDSSALNTTP 1005
Score = 33.5 bits (75), Expect = 0.13
Identities = 36/180 (20%), Positives = 69/180 (38%)
Frame = +1
Query: 117 VQLSDVSILSLQLQVKKLEDEKAKENEEKVETEPDLAVSGEGRLPENEKTGGDIDEPVPA 176
VQL+ S S + + +DE + + ++ D A SG+ +E + D
Sbjct: 1219 VQLTTTSDTSSEQK----KDESSSAESKSESSQNDNANSGQSNTTSDESANDNKDS---- 1374
Query: 177 IRRLDESTTNTDKLLPAAGEESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSG 236
+ TT+++ +A S+ +N S N + + + +G
Sbjct: 1375 ----SQVTTSSEN---SAEGNSNTENNSDENQNDSKNNENTNDSGNTSNDANVNENQNEN 1533
Query: 237 SVQVKSGLNEPDQTGRKRKSVEEESNNGSYDNNEAKAVTCESVPPSEERKVEDDSVTRSE 296
+ Q K+ NE D + ES +NNE+ ++ S ++ D SVT+ +
Sbjct: 1534 AAQTKTSENEGD--------AQNESVESKKENNESAHKDVDNNSNSNDQGSSDTSVTQDD 1689
>AW257087 homologue to GP|6683126|dbj KIAA0339 protein {Homo sapiens},
partial (2%)
Length = 466
Score = 35.8 bits (81), Expect = 0.027
Identities = 31/152 (20%), Positives = 58/152 (37%)
Frame = +1
Query: 193 AAGEESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLNEPDQTGR 252
++ S + S + + + S +A V V + P H + S + +
Sbjct: 4 SSSSSSSSSSSSSSSDSDSDSESEAPAKKAVPVVTKTAPTHAKSA---SSKMEVDSDSDS 174
Query: 253 KRKSVEEESNNGSYDNNEAKAVTCESVPPSEERKVEDDSVTRSEGGGTRESSEVQSSSSL 312
S S + S + + AV+ P+++ + + DS + + SS SSSS
Sbjct: 175 DSDSSSSSSTSSSVSSVKKPAVSAAKATPAKKEESDSDS----DSDSSSSSSSSSSSSSS 342
Query: 313 SKTRRTQRRRKEVSGEDAKMENEDVAVVKSEP 344
S + + + +D + E E VVK P
Sbjct: 343 SSSSSSSSSSSSSNSDDEEEEEEAKPVVKKTP 438
>TC81158 similar to GP|9280664|gb|AAF86533.1| F21B7.15 {Arabidopsis
thaliana}, partial (6%)
Length = 890
Score = 34.3 bits (77), Expect = 0.079
Identities = 22/62 (35%), Positives = 29/62 (46%)
Frame = +1
Query: 274 VTCESVPPSEERKVEDDSVTRSEGGGTRESSEVQSSSSLSKTRRTQRRRKEVSGEDAKME 333
+T PPS E +D SEG ESSE +S SS S + + E +DA+ E
Sbjct: 358 ITLTGDPPSVEVNGNEDKDDGSEGESESESSESESESSTSSSSTSGSDSDEDDEDDAEDE 537
Query: 334 NE 335
E
Sbjct: 538 EE 543
>TC79639 similar to GP|10728064|gb|AAF50455.2 CG7060 gene product
{Drosophila melanogaster}, partial (2%)
Length = 757
Score = 32.3 bits (72), Expect = 0.30
Identities = 19/93 (20%), Positives = 46/93 (49%)
Frame = +1
Query: 261 SNNGSYDNNEAKAVTCESVPPSEERKVEDDSVTRSEGGGTRESSEVQSSSSLSKTRRTQR 320
SNN +NN+ K ++ P+ + K ++ ++ +S+ + ++ ++ SK +
Sbjct: 85 SNNSVAENNKKKTKKPQTSTPTSKIKKDNKNIEKSKENNNKNKKKLNFTNGNSKEQEESG 264
Query: 321 RRKEVSGEDAKMENEDVAVVKSEPLFGVLEMIK 353
R+K+ + +D + E+ A P+ + M+K
Sbjct: 265 RKKKHAKDD---DEEEDAKTHVFPMNRIRTMLK 354
>TC84550 weakly similar to PIR|T46095|T46095 hypothetical protein T25B15.20
- Arabidopsis thaliana, partial (1%)
Length = 969
Score = 32.3 bits (72), Expect = 0.30
Identities = 23/86 (26%), Positives = 45/86 (51%), Gaps = 2/86 (2%)
Frame = +1
Query: 257 VEEESNNGSYDNNEAKAVTCESVPPSEERKVEDDSVTRSEGGGT--RESSEVQSSSSLSK 314
+ +++N G D ++A V SV +++ + D S+ T ES+ ++++S +K
Sbjct: 508 LNDDANGGESDTDDACVVEAGSVVDADKSGNKTDEDLPSDALNTFHDESNPLEATSLSAK 687
Query: 315 TRRTQRRRKEVSGEDAKMENEDVAVV 340
+ +E+SG + +EN DVA V
Sbjct: 688 LNES----REISGTEVCLENVDVASV 753
>TC89779 GP|12835923|dbj|BAB23419. data source:MGD source key:MGI:1333811
evidence:ISS~methyl-CpG binding domain protein
1~putative, partial (2%)
Length = 824
Score = 32.0 bits (71), Expect = 0.39
Identities = 21/55 (38%), Positives = 29/55 (52%), Gaps = 2/55 (3%)
Frame = +1
Query: 259 EESNNGSYDNNEAKAVTCESVP--PSEERKVEDDSVTRSEGGGTRESSEVQSSSS 311
EES++ D NE + ESVP SEE+KV++ S S G SE + +S
Sbjct: 379 EESDDDKRDYNERISEDRESVPQYSSEEKKVDESSERESVHGEEESESEGEQDNS 543
>BI308304 similar to GP|18642468|em transcriptional adaptor {Zea mays},
partial (24%)
Length = 781
Score = 31.6 bits (70), Expect = 0.51
Identities = 11/32 (34%), Positives = 21/32 (65%)
Frame = +2
Query: 21 WGTWEELLLASAVDRHGFKDWDTIAMEVQSRT 52
W EE+LL A+D +GF +W+ +A + +++
Sbjct: 536 WSAEEEMLLLEALDMYGFGNWNDVADNIGTKS 631
>TC81527 similar to GP|9954116|gb|AAG08961.1| tuber-specific and
sucrose-responsive element binding factor {Solanum
tuberosum}, partial (56%)
Length = 765
Score = 31.2 bits (69), Expect = 0.67
Identities = 13/49 (26%), Positives = 28/49 (56%), Gaps = 1/49 (2%)
Frame = +2
Query: 11 NKNPNQDLQG-WGTWEELLLASAVDRHGFKDWDTIAMEVQSRTNRTSLL 58
N+N ++G W E+ +L V++HG ++W I+ ++ R+ ++ L
Sbjct: 131 NQNKLDRIKGPWSAEEDRILTRLVEQHGARNWSLISRYIKGRSGKSCRL 277
>TC87287 similar to GP|7546725|gb|AAF63657.1| MFP1 attachment factor 1
{Lycopersicon esculentum}, partial (57%)
Length = 820
Score = 30.8 bits (68), Expect = 0.87
Identities = 36/153 (23%), Positives = 69/153 (44%), Gaps = 3/153 (1%)
Frame = +2
Query: 77 DDVPPPQQNGDVSAVTAEDSDHVPWLDKLRKQRVAELRRDVQLSDVSILSLQLQVKKLED 136
+ VPPPQQNGD + + P + R + L L+ S+LS + L
Sbjct: 230 NSVPPPQQNGDQTKPPSAGFSIWPPTQRTRDAVITRLIE--TLTTPSVLSKRYGT--LPS 397
Query: 137 EKAKENEEKVETEPDLAVSGEGRLPENEKTGGDIDEPVPAI-RRLDESTTNTDKLLPAAG 195
++A ++E E +V+G+ +P+ GGD E + + + + T K P
Sbjct: 398 DEAVSAARQIE-EDAFSVAGDSAVPD----GGDGIEILQVYSKEISKKMLETVKARPTVD 562
Query: 196 EES-DRDNQSVNES-NSTGSRFDALKTGEVDVK 226
+ D+D+ +V + + S DA ++ +++ +
Sbjct: 563 SSAVDKDSAAVAPTVDDPPSAEDAAESAKIETE 661
>TC85218 similar to SP|P05692|LEGJ_PEA Legumin J precursor. [Garden pea]
{Pisum sativum}, partial (73%)
Length = 1595
Score = 30.8 bits (68), Expect = 0.87
Identities = 27/109 (24%), Positives = 50/109 (45%), Gaps = 15/109 (13%)
Frame = +3
Query: 235 SGSVQVKSGLNEPDQTGRKRKSVEEESNNGSYD-NNEAKAVTCESVPPSE---------E 284
S S+++KS + G VEE++NN D N+ K C V
Sbjct: 318 SSSLKLKSDNKDDSNRGPVSPDVEEDANNKRRDLRNKTKVAVC*VVSAQSF*RRHSTRIR 497
Query: 285 RKVEDDSVTRSEGGGTRESSEVQSSSSLSKTRRTQR-----RRKEVSGE 328
++ D ++ + G + E V +SS+LS +++ + RRK+++G+
Sbjct: 498 IQLRDFNLHATRGAKS*E*RAVSASSALSGSKKMKNTKEVTRRKKMNGD 644
>TC85216 similar to PIR|S26688|S26688 legumin K - garden pea, partial (66%)
Length = 1582
Score = 30.8 bits (68), Expect = 0.87
Identities = 27/109 (24%), Positives = 50/109 (45%), Gaps = 15/109 (13%)
Frame = +3
Query: 235 SGSVQVKSGLNEPDQTGRKRKSVEEESNNGSYD-NNEAKAVTCESVPPSE---------E 284
S S+++KS + G VEE++NN D N+ K C V
Sbjct: 168 SSSLKLKSDNKDDSNRGPVSPDVEEDANNKRRDLRNKTKVAVC*VVSAQSF*RRHSTRIR 347
Query: 285 RKVEDDSVTRSEGGGTRESSEVQSSSSLSKTRRTQR-----RRKEVSGE 328
++ D ++ + G + E V +SS+LS +++ + RRK+++G+
Sbjct: 348 IQLRDFNLHATRGAKS*E*RAVSASSALSGSKKMKNTKEVTRRKKMNGD 494
>BQ140761 weakly similar to GP|18076245|emb phosphophoryn {Rattus
norvegicus}, partial (9%)
Length = 620
Score = 30.8 bits (68), Expect = 0.87
Identities = 29/119 (24%), Positives = 50/119 (41%), Gaps = 1/119 (0%)
Frame = +1
Query: 196 EESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQ-VKSGLNEPDQTGRKR 254
E+S + S + +S GS D+ + + D S S + V + E + +K
Sbjct: 238 EKSSDSSDSDSSDSSDGSDSDSDASDDSDDSSDSSSDDSSSDEEPVPAPKKEKAKKAKKA 417
Query: 255 KSVEEESNNGSYDNNEAKAVTCESVPPSEERKVEDDSVTRSEGGGTRESSEVQSSSSLS 313
KSV S++ ++E+++ E P + DDS + S + S SSS S
Sbjct: 418 KSVSSSSDSSDDSSSESES---EPEPXNVPLPESDDSSSASSDSDSSSDSSSDSSSDSS 585
>TC87590 weakly similar to PIR|G96522|G96522 F11A17.16 [imported] -
Arabidopsis thaliana, partial (45%)
Length = 2425
Score = 30.4 bits (67), Expect = 1.1
Identities = 25/89 (28%), Positives = 41/89 (45%), Gaps = 6/89 (6%)
Frame = +2
Query: 255 KSVEEE---SNNGSYDNNEAKAVTCESVPPSEERKVEDDSVTRSEGGGTRESSEVQSS-- 309
KS+EEE S+ GS + AK V V P R++E+D E E EV +
Sbjct: 545 KSIEEEVESSHKGSKEGEVAKVVV---VAPPRRRRIEEDDPDVKEKKELLEKLEVSENLI 715
Query: 310 -SSLSKTRRTQRRRKEVSGEDAKMENEDV 337
S S+ + +V G + +E++++
Sbjct: 716 KSLQSEINALKDELNQVKGLNIDLESQNI 802
>TC79702 MtN14
Length = 973
Score = 30.4 bits (67), Expect = 1.1
Identities = 29/108 (26%), Positives = 51/108 (46%), Gaps = 18/108 (16%)
Frame = +2
Query: 254 RKSVEEESNNGSYDNNEAKAVTCESV------PPSEERK----VEDDSVT----RSEGGG 299
++ VEE NG+ N E +SV P+EE + V+D++V +
Sbjct: 317 KEPVEETKENGNSLNVEETKENGDSVVEAVQEKPAEESETVNVVKDENVVAEPETKDNVK 496
Query: 300 TRESSEVQSSSSLSKTRRTQRRRKE--VSGEDAKME--NEDVAVVKSE 343
T E+SE ++ + K +++E ++ DAK+E E+ + KSE
Sbjct: 497 TEETSEEKNEEKVEKEDAMDEKKEEEVITNNDAKIEKNEEEAPIEKSE 640
>TC86730 similar to GP|13811646|gb|AAK40224.1 syntaxin of plants 52
{Arabidopsis thaliana}, partial (98%)
Length = 1279
Score = 30.0 bits (66), Expect = 1.5
Identities = 19/62 (30%), Positives = 33/62 (52%)
Frame = +1
Query: 113 LRRDVQLSDVSILSLQLQVKKLEDEKAKENEEKVETEPDLAVSGEGRLPENEKTGGDIDE 172
+ R V L + ++ LQ QV K +DE ++ EE V + +A++ L + + D+DE
Sbjct: 634 MSRTVGLDNNGLVGLQRQVMKEQDEGLEKLEETVLSTKHIALAVNEELTLHTRLIDDLDE 813
Query: 173 PV 174
V
Sbjct: 814 HV 819
>TC87918 weakly similar to GP|15810397|gb|AAL07086.1 unknown protein
{Arabidopsis thaliana}, partial (67%)
Length = 1746
Score = 30.0 bits (66), Expect = 1.5
Identities = 25/91 (27%), Positives = 40/91 (43%), Gaps = 18/91 (19%)
Frame = +2
Query: 181 DESTTNTDKLLPA--AGEESD----------RDNQSVNESNST-GSRFDALKTG-----E 222
+ T TD+ P+ A EESD ++N VNE G + K G E
Sbjct: 830 NNDVTKTDEQKPSEVAAEESDQKMSENDDNDKENNGVNEGKEAEGEEKEQEKNGVKEEKE 1009
Query: 223 VDVKLGSGPGHGSGSVQVKSGLNEPDQTGRK 253
+V+ +G+G +V G+ + +TG+K
Sbjct: 1010TEVEEKGQENNGAGEGKVTEGVEKGQETGKK 1102
>TC87391 homologue to GP|22832155|gb|AAF48195.2 CG4013-PA {Drosophila
melanogaster}, partial (0%)
Length = 2359
Score = 29.6 bits (65), Expect = 1.9
Identities = 70/304 (23%), Positives = 114/304 (37%), Gaps = 40/304 (13%)
Frame = +3
Query: 51 RTNRTSLLATAHHCEQKF---HDLNRR-FKDDVPPPQQNGDVSAVTAEDSDHVPWLDKLR 106
+ R++ L C+QK H +R FK P + + AV DSD V D
Sbjct: 1521 KNRRSTRLHNIKVCQQKKPKQHPESRGPFKH--PDGEPSSTTMAVENHDSDSVVPDDV-- 1688
Query: 107 KQRVAELRRDVQLSDV--SILSLQLQVKKLEDEKAKENEEKVETEPDLAVSGEGRL---- 160
+ E+ D + V S+++ + LEDE+ + ++ V G GRL
Sbjct: 1689 PANLPEISFDALENSVPTSLVTTDCNMSFLEDEQ----DMSIKMSEVNNVHGSGRLHNTK 1856
Query: 161 ---------------PENEKTGG-------DIDEPVPAIRRLDESTTNTDKLLPAAGEES 198
PE E++ D+D VP D +TN ++L A E+S
Sbjct: 1857 DCQKKQPASGDPLKNPEKEQSSTLKVVENHDLDTAVP-----DNVSTNLPEILSDAVEKS 2021
Query: 199 --------DRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLNEPDQT 250
D+ N++ R ++KTG V+V HGSG + + ++
Sbjct: 2022 FPTSLVSLDKTTGDRNKAFLEDERDMSIKTGRVNVV------HGSGGNKRSARMHNVKPY 2183
Query: 251 GRKRKSVEEESNNGSYDNNEAKAVTCESVPPSEERKVEDDSVTRSEGGGTRESSEVQSSS 310
+K+ + + + + D A AV + S+GG S+ VQS
Sbjct: 2184 QKKQPATGDPATHPDKDQISASAV-----------------LKMSDGGLGSLSTPVQSQK 2312
Query: 311 SLSK 314
SK
Sbjct: 2313 GKSK 2324
>TC82818 similar to GP|20259203|gb|AAM14317.1 unknown protein {Arabidopsis
thaliana}, partial (43%)
Length = 1176
Score = 29.3 bits (64), Expect = 2.5
Identities = 14/36 (38%), Positives = 20/36 (54%)
Frame = +1
Query: 41 WDTIAMEVQSRTNRTSLLATAHHCEQKFHDLNRRFK 76
W +I+ + R R S CE KF+DLN+R+K
Sbjct: 7 WKSISKVMAERGYRVS----PQQCEDKFNDLNKRYK 102
>TC82954 weakly similar to GP|8809591|dbj|BAA97142.1
emb|CAB82814.1~gene_id:MNB8.8~similar to unknown protein
{Arabidopsis thaliana}, partial (7%)
Length = 783
Score = 29.3 bits (64), Expect = 2.5
Identities = 33/109 (30%), Positives = 49/109 (44%), Gaps = 16/109 (14%)
Frame = +3
Query: 105 LRKQRVAELRRDVQLSDVSILSLQLQ------VKKLEDE--KAKENEEKV-ETEPDLAVS 155
L + RV E +RD+ +SD++ +++ KK+E+E K KE EEK + E D
Sbjct: 441 LNELRVFEKKRDLAISDLNQKLKEMEGLVEEKDKKIEEEEKKRKELEEKAKKAEKDAEEL 620
Query: 156 GEGRLPENEKTGGDID-------EPVPAIRRLDESTTNTDKLLPAAGEE 197
E E ++ D+ E V R L+ K L AA EE
Sbjct: 621 RESSKREGQEHSSDLRKHKTAFIELVSNQRHLEAELGRAVKHLDAAKEE 767
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.306 0.126 0.345
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,350,043
Number of Sequences: 36976
Number of extensions: 110033
Number of successful extensions: 546
Number of sequences better than 10.0: 62
Number of HSP's better than 10.0 without gapping: 533
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 544
length of query: 371
length of database: 9,014,727
effective HSP length: 98
effective length of query: 273
effective length of database: 5,391,079
effective search space: 1471764567
effective search space used: 1471764567
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (22.0 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146756.1


