

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146755.12 + phase: 0
(57 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
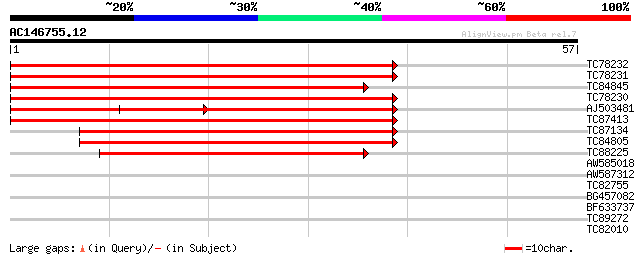
Score E
Sequences producing significant alignments: (bits) Value
TC78232 similar to GP|21593805|gb|AAM65772.1 putativepod-specifi... 78 6e-16
TC78231 similar to GP|21593805|gb|AAM65772.1 putativepod-specifi... 74 1e-14
TC84845 similar to PIR|T10561|T10561 hypothetical protein F25E4.... 71 1e-13
TC78230 similar to PIR|T10561|T10561 hypothetical protein F25E4.... 70 2e-13
AJ503481 similar to PIR|T05380|T053 hypothetical protein F16G20.... 44 2e-10
TC87413 similar to PIR|T10561|T10561 hypothetical protein F25E4.... 54 2e-08
TC87134 similar to PIR|T48275|T48275 hypothetical protein T22P11... 44 1e-05
TC84805 similar to GP|9663977|dbj|BAB03618.1 contains ESTs AU064... 44 1e-05
TC88225 similar to GP|21593805|gb|AAM65772.1 putativepod-specifi... 41 8e-05
AW585018 similar to GP|15146202|gb AT5g50130/MPF21_15 {Arabidops... 33 0.029
AW587312 similar to GP|15081624|gb| At1g64590/F1N19_15 {Arabidop... 30 0.15
TC82755 25 4.7
BG457082 similar to PIR|T05452|T054 hypothetical protein F7K2.16... 25 6.1
BF633737 similar to GP|23498351|emb hypothetical protein conser... 25 6.1
TC89272 similar to GP|11994729|dbj|BAB03045. gene_id:F5N5.17~pir... 25 6.1
TC82010 similar to GP|15293217|gb|AAK93719.1 unknown protein {Ar... 25 8.0
>TC78232 similar to GP|21593805|gb|AAM65772.1 putativepod-specific
dehydrogenase SAC25 {Arabidopsis thaliana}, partial
(78%)
Length = 914
Score = 78.2 bits (191), Expect = 6e-16
Identities = 37/39 (94%), Positives = 38/39 (96%)
Frame = +2
Query: 1 MWPFSKKGVSGFSGNSTAEKVTHGIDASGLTAIVTGLSS 39
MWPFSKKGVSGFSGNSTAEKVTHGIDASGLTAIVTG +S
Sbjct: 68 MWPFSKKGVSGFSGNSTAEKVTHGIDASGLTAIVTGATS 184
>TC78231 similar to GP|21593805|gb|AAM65772.1 putativepod-specific
dehydrogenase SAC25 {Arabidopsis thaliana}, partial
(82%)
Length = 1318
Score = 73.6 bits (179), Expect = 1e-14
Identities = 35/39 (89%), Positives = 37/39 (94%)
Frame = +3
Query: 1 MWPFSKKGVSGFSGNSTAEKVTHGIDASGLTAIVTGLSS 39
MWPFSKKGVSGFS NSTAE+VTHGIDA+GLTAIVTG SS
Sbjct: 60 MWPFSKKGVSGFSWNSTAEQVTHGIDATGLTAIVTGASS 176
>TC84845 similar to PIR|T10561|T10561 hypothetical protein F25E4.30 -
Arabidopsis thaliana, partial (11%)
Length = 672
Score = 70.9 bits (172), Expect = 1e-13
Identities = 33/36 (91%), Positives = 35/36 (96%)
Frame = +3
Query: 1 MWPFSKKGVSGFSGNSTAEKVTHGIDASGLTAIVTG 36
MWPFSKKGVSGFS NSTAE+VTHGIDA+GLTAIVTG
Sbjct: 54 MWPFSKKGVSGFSWNSTAEQVTHGIDATGLTAIVTG 161
>TC78230 similar to PIR|T10561|T10561 hypothetical protein F25E4.30 -
Arabidopsis thaliana, partial (90%)
Length = 1121
Score = 70.1 bits (170), Expect = 2e-13
Identities = 33/39 (84%), Positives = 36/39 (91%)
Frame = +2
Query: 1 MWPFSKKGVSGFSGNSTAEKVTHGIDASGLTAIVTGLSS 39
MWPFS+KGVSGFS STAE+VTHGIDA+GLTAIVTG SS
Sbjct: 50 MWPFSRKGVSGFSWKSTAEEVTHGIDATGLTAIVTGASS 166
>AJ503481 similar to PIR|T05380|T053 hypothetical protein F16G20.120 -
Arabidopsis thaliana, partial (46%)
Length = 343
Score = 43.5 bits (101), Expect(2) = 2e-10
Identities = 22/28 (78%), Positives = 23/28 (81%)
Frame = +2
Query: 12 FSGNSTAEKVTHGIDASGLTAIVTGLSS 39
F STAE+VTHGIDA GLTAIVTG SS
Sbjct: 86 FLRESTAEQVTHGIDAIGLTAIVTGASS 169
Score = 36.2 bits (82), Expect(2) = 2e-10
Identities = 14/20 (70%), Positives = 16/20 (80%)
Frame = +3
Query: 1 MWPFSKKGVSGFSGNSTAEK 20
MWPFSKKG+SGFSG+ K
Sbjct: 54 MWPFSKKGISGFSGSQLLNK 113
>TC87413 similar to PIR|T10561|T10561 hypothetical protein F25E4.30 -
Arabidopsis thaliana, partial (94%)
Length = 1192
Score = 53.5 bits (127), Expect = 2e-08
Identities = 25/39 (64%), Positives = 30/39 (76%)
Frame = +1
Query: 1 MWPFSKKGVSGFSGNSTAEKVTHGIDASGLTAIVTGLSS 39
MW KG SGFS +STA++VTHGID + LTAI+TG SS
Sbjct: 67 MWFIGWKGPSGFSASSTAQQVTHGIDGTSLTAIITGASS 183
>TC87134 similar to PIR|T48275|T48275 hypothetical protein T22P11.130 -
Arabidopsis thaliana, partial (87%)
Length = 1374
Score = 43.9 bits (102), Expect = 1e-05
Identities = 21/32 (65%), Positives = 26/32 (80%)
Frame = +3
Query: 8 GVSGFSGNSTAEKVTHGIDASGLTAIVTGLSS 39
G SGF ++TAE+VT GIDAS LTAI+TG +S
Sbjct: 270 GPSGFGSSTTAEQVTQGIDASNLTAIITGGAS 365
>TC84805 similar to GP|9663977|dbj|BAB03618.1 contains ESTs AU064282(E20459)
AU097686(S6509)~unknown protein, partial (47%)
Length = 634
Score = 43.9 bits (102), Expect = 1e-05
Identities = 22/32 (68%), Positives = 25/32 (77%)
Frame = +3
Query: 8 GVSGFSGNSTAEKVTHGIDASGLTAIVTGLSS 39
G SGF STAE+VT GIDAS LTAI+TG +S
Sbjct: 81 GPSGFGSASTAEQVTQGIDASNLTAIITGGAS 176
>TC88225 similar to GP|21593805|gb|AAM65772.1 putativepod-specific
dehydrogenase SAC25 {Arabidopsis thaliana}, partial
(67%)
Length = 821
Score = 41.2 bits (95), Expect = 8e-05
Identities = 20/27 (74%), Positives = 23/27 (85%)
Frame = +3
Query: 10 SGFSGNSTAEKVTHGIDASGLTAIVTG 36
S F+ +STAE+VTHGID SGL AIVTG
Sbjct: 111 SEFTLSSTAEEVTHGIDGSGLAAIVTG 191
>AW585018 similar to GP|15146202|gb AT5g50130/MPF21_15 {Arabidopsis
thaliana}, partial (63%)
Length = 705
Score = 32.7 bits (73), Expect = 0.029
Identities = 18/35 (51%), Positives = 22/35 (62%), Gaps = 3/35 (8%)
Frame = +3
Query: 8 GVSGFSGNSTAEKVTHGID---ASGLTAIVTGLSS 39
G SGF NSTAE+VT S LTA++TG +S
Sbjct: 66 GPSGFGSNSTAEQVTQHCSLFIPSNLTALITGATS 170
>AW587312 similar to GP|15081624|gb| At1g64590/F1N19_15 {Arabidopsis
thaliana}, partial (27%)
Length = 628
Score = 30.4 bits (67), Expect = 0.15
Identities = 17/33 (51%), Positives = 21/33 (63%), Gaps = 1/33 (3%)
Frame = +2
Query: 8 GVSGFSGNSTAEKVTHGI-DASGLTAIVTGLSS 39
G SGF STAE+VT D +TAI+TG +S
Sbjct: 374 GPSGFGSKSTAEQVTETCGDLRSITAIITGGTS 472
>TC82755
Length = 661
Score = 25.4 bits (54), Expect = 4.7
Identities = 11/18 (61%), Positives = 13/18 (72%)
Frame = +2
Query: 37 LSSITLSYHFFNFKIFKH 54
LSSI S HFF F ++KH
Sbjct: 11 LSSILQSLHFFFFFLYKH 64
>BG457082 similar to PIR|T05452|T054 hypothetical protein F7K2.160 -
Arabidopsis thaliana, partial (4%)
Length = 623
Score = 25.0 bits (53), Expect = 6.1
Identities = 10/15 (66%), Positives = 12/15 (79%)
Frame = -1
Query: 6 KKGVSGFSGNSTAEK 20
KKG+ GF G+S AEK
Sbjct: 101 KKGIKGFLGDSEAEK 57
>BF633737 similar to GP|23498351|emb hypothetical protein conserved
{Plasmodium falciparum 3D7}, partial (2%)
Length = 560
Score = 25.0 bits (53), Expect = 6.1
Identities = 10/15 (66%), Positives = 12/15 (79%)
Frame = -2
Query: 6 KKGVSGFSGNSTAEK 20
KKG+ GF G+S AEK
Sbjct: 127 KKGIKGFLGDSEAEK 83
>TC89272 similar to GP|11994729|dbj|BAB03045.
gene_id:F5N5.17~pir||G71408~similar to unknown protein
{Arabidopsis thaliana}, partial (57%)
Length = 1366
Score = 25.0 bits (53), Expect = 6.1
Identities = 18/48 (37%), Positives = 24/48 (49%), Gaps = 8/48 (16%)
Frame = -1
Query: 13 SGNSTAEKVTH--------GIDASGLTAIVTGLSSITLSYHFFNFKIF 52
S +TA+ VT G ++ G +I T SS LS F+FKIF
Sbjct: 1171 SSKTTAKPVTTLTPVNEPLGFNSGGFHSITTFSSSPELSVTQFSFKIF 1028
>TC82010 similar to GP|15293217|gb|AAK93719.1 unknown protein {Arabidopsis
thaliana}, partial (62%)
Length = 1304
Score = 24.6 bits (52), Expect = 8.0
Identities = 12/21 (57%), Positives = 14/21 (66%)
Frame = +2
Query: 16 STAEKVTHGIDASGLTAIVTG 36
+T EKV + ASG AIVTG
Sbjct: 1229 ATTEKVQQYVKASGKEAIVTG 1291
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.135 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,785,456
Number of Sequences: 36976
Number of extensions: 15683
Number of successful extensions: 123
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 123
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 123
length of query: 57
length of database: 9,014,727
effective HSP length: 33
effective length of query: 24
effective length of database: 7,794,519
effective search space: 187068456
effective search space used: 187068456
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 51 (24.3 bits)
Medicago: description of AC146755.12


