

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146752.6 + phase: 0
(346 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
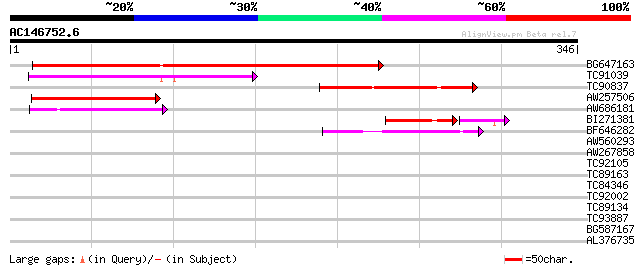
Score E
Sequences producing significant alignments: (bits) Value
BG647163 similar to GP|9757943|dbj emb|CAB72466.1~gene_id:MJC20.... 206 1e-53
TC91039 similar to PIR|C96640|C96640 hypothetical protein T25B24... 122 2e-28
TC90837 weakly similar to GP|22202752|dbj|BAC07409. hypothetical... 91 9e-19
AW257506 weakly similar to GP|9757943|dbj| emb|CAB72466.1~gene_i... 65 4e-11
AW686181 58 6e-09
BI271381 similar to GP|21397269|gb Unknown protein {Oryza sativa... 46 2e-06
BF646282 weakly similar to GP|17104745|gb| unknown protein {Arab... 42 3e-04
AW560293 weakly similar to GP|21902022|db contains EST D24834(R2... 34 0.073
AW267858 weakly similar to GP|21592524|gb unknown {Arabidopsis t... 33 0.12
TC92105 homologue to GP|16197625|gb|AAL13436.1 anaphase promotin... 32 0.36
TC89163 similar to GP|12044358|gb|AAG47821.1 SEX1 {Arabidopsis t... 32 0.47
TC84346 similar to PIR|D85062|D85062 hypothetical protein AT4g04... 30 1.4
TC92002 similar to GP|4104929|gb|AAD02218.1| auxin response fact... 28 5.2
TC89134 weakly similar to GP|13430834|gb|AAK26039.1 unknown prot... 28 5.2
TC93887 similar to GP|19335722|gb|AAL85485.1 transporter associa... 28 6.8
BG587167 similar to PIR|T47439|T474 hypothetical protein T18B22.... 27 8.9
AL376735 similar to GP|870794|gb|AA polyubiquitin {Arabidopsis t... 27 8.9
>BG647163 similar to GP|9757943|dbj emb|CAB72466.1~gene_id:MJC20.8~similar to
unknown protein {Arabidopsis thaliana}, partial (51%)
Length = 744
Score = 206 bits (523), Expect = 1e-53
Identities = 105/214 (49%), Positives = 138/214 (64%)
Frame = +3
Query: 15 RMGPQTFLKLCDMLEREGGLRPTRWSSVEEQVAKSLYILTHNAKNREVNFWFRRSGETIS 74
RM F KLCD+LE +G LR T +EEQ+A L+I+ HN + R V F SGETIS
Sbjct: 93 RMDKVVFYKLCDILETKGLLRDTNRIKIEEQLAIFLFIIGHNLRIRGVQELFHYSGETIS 272
Query: 75 RHLHQVLKAILELEEKFIVQPDGSTIPLEISSSTRFYPYFKDCVGAIDGTHIRVKVSAKD 134
RH + VL A++ + +++ QP G + I+ RF+PYFKDCVGAIDG ++ V V +
Sbjct: 273 RHFNNVLNAVMSISKEYF-QPPGEDVASMIAEDDRFFPYFKDCVGAIDGIYVPVTVGVDE 449
Query: 135 APRYRGRKEYPTQNVLAACTFDLKFTYVLAGWEGSASDSRIIKNALTREDKLKIPEGKYY 194
+R + +QNVLAAC+FDLKF YVLAGWEGSAS+ ++ +A+TR++KL++PEGKYY
Sbjct: 450 QGPFRNKDGLLSQNVLAACSFDLKFCYVLAGWEGSASNLQVFNSAITRKNKLQVPEGKYY 629
Query: 195 LVDAGFMLTSGLITPYRGVRYHLKEFSARNPPLN 228
LVD F G I PY YH KEF P N
Sbjct: 630 LVDNKFPNVPGFIAPYPRTPYHSKEFPTGYQPQN 731
>TC91039 similar to PIR|C96640|C96640 hypothetical protein T25B24.14
[imported] - Arabidopsis thaliana, partial (4%)
Length = 732
Score = 122 bits (307), Expect = 2e-28
Identities = 68/149 (45%), Positives = 90/149 (59%), Gaps = 9/149 (6%)
Frame = +1
Query: 12 NIIRMGPQTFLKLCDMLEREGGLRPTRWSSVEEQVAKSLYILTHNAKNREVNFWFRRSGE 71
N RM P F +LC+ L+ + GL+P++ +VEE+V +Y L A NR+V F+ SGE
Sbjct: 283 NAFRMDPTLFKQLCEDLQSKYGLQPSKRMTVEEKVGIFVYTLAMGASNRDVRERFQHSGE 462
Query: 72 TISRHLHQVLKAILELEEKF------IVQPDGST---IPLEISSSTRFYPYFKDCVGAID 122
TISR H+VL+AI + I++P T IPL IS+ R+ PYFKDC+G ID
Sbjct: 463 TISRAFHEVLEAISGRSRGYRGLARDIIRPKDPTFQFIPLHISNDERYMPYFKDCIGCID 642
Query: 123 GTHIRVKVSAKDAPRYRGRKEYPTQNVLA 151
GTHI + D RYRGRK PT NV+A
Sbjct: 643 GTHIAACIPEADQMRYRGRKGIPTFNVMA 729
>TC90837 weakly similar to GP|22202752|dbj|BAC07409. hypothetical
protein~similar to Oryza sativa chromosome10
OSJNBa0079H13.17, partial (7%)
Length = 596
Score = 90.5 bits (223), Expect = 9e-19
Identities = 45/96 (46%), Positives = 62/96 (63%)
Frame = +1
Query: 190 EGKYYLVDAGFMLTSGLITPYRGVRYHLKEFSARNPPLNYKELFNLRHASLRNAIERAFG 249
+G+YYLVD G+ G + PY +RYH +F PP N +E FN H+SLR+ IER+FG
Sbjct: 1 DGRYYLVDKGYPDKEGYMVPYPRIRYHQSQFE-HEPPTNAQEAFNRAHSSLRSCIERSFG 177
Query: 250 VLKKRFEILSNSTEPAYGIKAQKLIIFACCILHNYL 285
VLKKR++IL+ P + +K Q +I A LHNY+
Sbjct: 178 VLKKRWKILNKM--PQFSVKTQIDVIIAAFALHNYI 279
>AW257506 weakly similar to GP|9757943|dbj|
emb|CAB72466.1~gene_id:MJC20.8~similar to unknown
protein {Arabidopsis thaliana}, partial (9%)
Length = 510
Score = 65.1 bits (157), Expect = 4e-11
Identities = 34/79 (43%), Positives = 51/79 (64%)
Frame = +2
Query: 14 IRMGPQTFLKLCDMLEREGGLRPTRWSSVEEQVAKSLYILTHNAKNREVNFWFRRSGETI 73
+R+ +TF KLC +L+ +G L TR E VA L+IL HN K R V+F + RS ETI
Sbjct: 218 LRVSKRTFFKLCKILKEKGQLVRTRNVPTTEAVAMFLHILAHNLKYRVVHFSYCRSKETI 397
Query: 74 SRHLHQVLKAILELEEKFI 92
SR + VL+AI+++ +++
Sbjct: 398 SRQFNNVLRAIMKVSREYL 454
>AW686181
Length = 304
Score = 57.8 bits (138), Expect = 6e-09
Identities = 34/84 (40%), Positives = 47/84 (55%)
Frame = +3
Query: 13 IIRMGPQTFLKLCDMLEREGGLRPTRWSSVEEQVAKSLYILTHNAKNREVNFWFRRSGET 72
+ RM F KLC L E L+ ++ VEE VA L ++ H NR + F+ SGET
Sbjct: 6 MFRMEKHIFHKLCHELV-EHDLKSSKHMGVEEMVAMFLVVVGHGVGNRMIQERFQHSGET 182
Query: 73 ISRHLHQVLKAILELEEKFIVQPD 96
+SRH ++VL A L+L K+I D
Sbjct: 183VSRHFYRVLHACLKLSFKYIKPED 254
>BI271381 similar to GP|21397269|gb Unknown protein {Oryza sativa (japonica
cultivar-group)}, partial (2%)
Length = 407
Score = 45.8 bits (107), Expect(2) = 2e-06
Identities = 25/44 (56%), Positives = 29/44 (65%)
Frame = +2
Query: 230 KELFNLRHASLRNAIERAFGVLKKRFEILSNSTEPAYGIKAQKL 273
+ELFN RH+SLR IER FGVLK RF IL P+Y Q+L
Sbjct: 137 EELFNYRHSSLRMTIERCFGVLKNRFPIL--KLMPSYKPSRQRL 262
Score = 23.5 bits (49), Expect(2) = 2e-06
Identities = 10/37 (27%), Positives = 19/37 (51%), Gaps = 6/37 (16%)
Frame = +3
Query: 275 IFACCILHNYLMSAEPNEDL------IAEVDAELANQ 305
+ ACC +HNY+ ++L + +++ E NQ
Sbjct: 264 VTACCAIHNYICKWNLPDELFRIWEEMDDIELEAVNQ 374
>BF646282 weakly similar to GP|17104745|gb| unknown protein {Arabidopsis
thaliana}, partial (15%)
Length = 570
Score = 42.0 bits (97), Expect = 3e-04
Identities = 29/98 (29%), Positives = 46/98 (46%)
Frame = +1
Query: 192 KYYLVDAGFMLTSGLITPYRGVRYHLKEFSARNPPLNYKELFNLRHASLRNAIERAFGVL 251
+Y + D GF L L+TPY G N + FN R A+ + +RA L
Sbjct: 1 EYIVGDTGFPLLPWLLTPYEGEGLS-----------NVQVDFNKRVAATQMVAKRALSRL 147
Query: 252 KKRFEILSNSTEPAYGIKAQKLIIFACCILHNYLMSAE 289
K+ ++I+ + K ++I+ CCILHN ++ E
Sbjct: 148 KEMWKIIGGVMQKPNKHKLPRIIL-VCCILHNIVIHLE 258
>AW560293 weakly similar to GP|21902022|db contains EST D24834(R2633)~similar
to Arabidopsis thaliana chromosome 5 At5g12010~unknown
protein, partial (25%)
Length = 623
Score = 34.3 bits (77), Expect = 0.073
Identities = 34/162 (20%), Positives = 69/162 (41%), Gaps = 19/162 (11%)
Frame = -2
Query: 168 GSASDSRII-KNALTREDKLKIPEGKYYLVDAGFMLTSGLITPYRGVRYHLKEFSARNPP 226
GS +D +++ K+ + + + + + ++GF L G++ PY + +
Sbjct: 622 GSYNDDQVLEKSVMYQRAMTGNLKDSWVVGNSGFPLMDGILVPYTHQNLTWTQHA----- 458
Query: 227 LNYKELFNLRHASLRNAIERAFGVLKKRFEILSNSTEPAYGIKAQKL--IIFACCILHNY 284
FN + ++ + AF +K R+ L E +K ++L ++ ACC+LHN
Sbjct: 457 ------FNEKVEDIQKLSKDAFAKVKGRWSCLQKRIE----VKIEELPGVLGACCVLHNI 308
Query: 285 LMSAEP-----------NEDLIAE-----VDAELANQNVSHD 310
+++++AE V A A N++HD
Sbjct: 307 CEMRNEKMDPAWNFELFDDEMVAENGVKSVAAAQARDNIAHD 182
>AW267858 weakly similar to GP|21592524|gb unknown {Arabidopsis thaliana},
partial (21%)
Length = 556
Score = 33.5 bits (75), Expect = 0.12
Identities = 29/110 (26%), Positives = 50/110 (45%), Gaps = 4/110 (3%)
Frame = +1
Query: 193 YYLVDAGFMLTSGLITPYRGVRYHLKEFSARNPPLNYKELFNLRHASLRNAIERAFGVLK 252
+ ++++G+ L ++ PY+ + FN R + + AF LK
Sbjct: 46 WIVLNSGYSLMDWVLVPYKHQNLTWTQHG-----------FNERIGEIEKIGKDAFSRLK 192
Query: 253 KRFEILSNSTEPAYGIKAQKL--IIFACCILHNY--LMSAEPNEDLIAEV 298
R+ L TE IK Q L ++ ACC+LHN +M+ E +++ EV
Sbjct: 193 GRWGCLKKRTE----IKLQDLPVVLGACCVLHNICEIMNEEMDDEWKFEV 330
>TC92105 homologue to GP|16197625|gb|AAL13436.1 anaphase promoting complex
subunit 11 {Arabidopsis thaliana}, partial (98%)
Length = 549
Score = 32.0 bits (71), Expect = 0.36
Identities = 11/21 (52%), Positives = 15/21 (71%)
Frame = +3
Query: 152 ACTFDLKFTYVLAGWEGSASD 172
AC+FD +F Y L+GW+ S D
Sbjct: 462 ACSFDFEFMYALSGWKSSTHD 524
>TC89163 similar to GP|12044358|gb|AAG47821.1 SEX1 {Arabidopsis thaliana},
partial (26%)
Length = 1158
Score = 31.6 bits (70), Expect = 0.47
Identities = 18/58 (31%), Positives = 31/58 (53%)
Frame = +2
Query: 238 ASLRNAIERAFGVLKKRFEILSNSTEPAYGIKAQKLIIFACCILHNYLMSAEPNEDLI 295
+++R A+ER + L N+ P +K++ F C +L N +S++ NEDLI
Sbjct: 701 SAVRTAVERGYEEL--------NNAGP------EKIMYFICLVLENLALSSDDNEDLI 832
>TC84346 similar to PIR|D85062|D85062 hypothetical protein AT4g04960
[imported] - Arabidopsis thaliana, partial (34%)
Length = 844
Score = 30.0 bits (66), Expect = 1.4
Identities = 14/49 (28%), Positives = 27/49 (54%), Gaps = 3/49 (6%)
Frame = +2
Query: 298 VDAELANQNVSHDNH---EASRSDMDEFAQGGIIKNGAAHQMWSNYKNN 343
+D N VSHD + +S+ D+ + ++ NG +Q+W +YK++
Sbjct: 455 IDINSLNSVVSHDVGFWVDDEKSEKDQIFEKLVLNNGENYQVWIDYKDS 601
>TC92002 similar to GP|4104929|gb|AAD02218.1| auxin response factor 7
{Arabidopsis thaliana}, partial (31%)
Length = 1138
Score = 28.1 bits (61), Expect = 5.2
Identities = 21/57 (36%), Positives = 29/57 (50%), Gaps = 1/57 (1%)
Frame = +3
Query: 264 PAYGIKAQKLIIFACCILHNYLMSAEPNEDLIAEVDAELANQNVS-HDNHEASRSDM 319
P+Y KLI C+LHN + A+P D EV A++ Q V+ +D SDM
Sbjct: 12 PSYPNLPSKLI----CMLHNVALHADPETD---EVYAQMTLQPVNKYDKEAMLASDM 161
>TC89134 weakly similar to GP|13430834|gb|AAK26039.1 unknown protein
{Arabidopsis thaliana}, partial (48%)
Length = 1122
Score = 28.1 bits (61), Expect = 5.2
Identities = 18/52 (34%), Positives = 26/52 (49%), Gaps = 6/52 (11%)
Frame = -1
Query: 101 PLEISSST------RFYPYFKDCVGAIDGTHIRVKVSAKDAPRYRGRKEYPT 146
P+E S+T +F PYFK C+G + H ++ V P+Y RK T
Sbjct: 258 PVEDISNTPVPP*AKFIPYFKSCLGTVLVFH-KL*VIGDFNPKYLARKSVKT 106
>TC93887 similar to GP|19335722|gb|AAL85485.1 transporter associated with
antigen processing-like protein {Arabidopsis thaliana},
partial (15%)
Length = 739
Score = 27.7 bits (60), Expect = 6.8
Identities = 14/52 (26%), Positives = 27/52 (51%)
Frame = +3
Query: 271 QKLIIFACCILHNYLMSAEPNEDLIAEVDAELANQNVSHDNHEASRSDMDEF 322
Q LI F+ H++ +S+ L + + + N+ H+N E++ + DEF
Sbjct: 150 QPLITFSSVNRHHFTLSSSKRILLSSSIKSSSINEVSIHNNSESASNASDEF 305
>BG587167 similar to PIR|T47439|T474 hypothetical protein T18B22.40 -
Arabidopsis thaliana, partial (11%)
Length = 788
Score = 27.3 bits (59), Expect = 8.9
Identities = 13/38 (34%), Positives = 20/38 (52%)
Frame = +2
Query: 264 PAYGIKAQKLIIFACCILHNYLMSAEPNEDLIAEVDAE 301
P + K +K I+ AC LHN+L ++ E D+E
Sbjct: 545 PPFPYKTKKEIVLACVGLHNFLRKFCRTDNFPEEKDSE 658
>AL376735 similar to GP|870794|gb|AA polyubiquitin {Arabidopsis thaliana},
partial (4%)
Length = 400
Score = 27.3 bits (59), Expect = 8.9
Identities = 18/66 (27%), Positives = 34/66 (51%), Gaps = 3/66 (4%)
Frame = +2
Query: 72 TISRHLHQVLKAILELEEKFIVQPDGSTIPLEISSSTRFYP---YFKDCVGAIDGTHIRV 128
T ++++ +V + L KF+V P G + PLEI R +P + + +DG + +
Sbjct: 173 TFAKNVQRV--TLKTLSSKFLVIPFGFSEPLEIEICIRSFPCDSMMQIFIKKLDGKVLPL 346
Query: 129 KVSAKD 134
+V + D
Sbjct: 347 RVKSSD 364
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.135 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,246,867
Number of Sequences: 36976
Number of extensions: 129827
Number of successful extensions: 619
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 613
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 615
length of query: 346
length of database: 9,014,727
effective HSP length: 97
effective length of query: 249
effective length of database: 5,428,055
effective search space: 1351585695
effective search space used: 1351585695
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146752.6


