

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
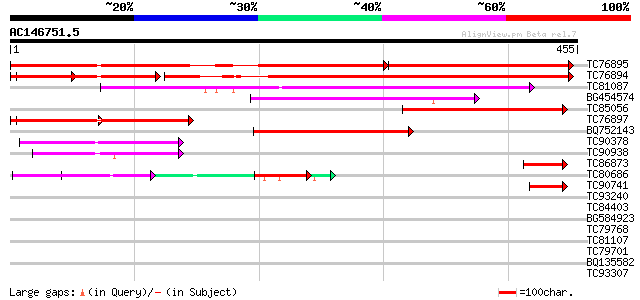
Query= AC146751.5 + phase: 0 /pseudo
(455 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC76895 homologue to SP|P35100|CLPA_PEA ATP-dependent clp protea... 244 e-110
TC76894 homologue to SP|P35100|CLPA_PEA ATP-dependent clp protea... 300 e-109
TC81087 homologue to GP|530207|gb|AAA66338.1|| heat shock protei... 197 7e-51
BG454574 similar to SP|P42762|ERD1 ERD1 protein chloroplast pre... 140 1e-33
TC85056 homologue to GP|9651530|gb|AAF91178.1| ClpB {Phaseolus l... 133 1e-31
TC76897 homologue to SP|P35100|CLPA_PEA ATP-dependent clp protea... 128 5e-30
BQ752143 similar to PIR|T39572|T39 probable proteinase subunit -... 112 2e-25
TC90378 weakly similar to GP|21592785|gb|AAM64734.1 unknown {Ara... 50 2e-06
TC90938 similar to GP|21592785|gb|AAM64734.1 unknown {Arabidopsi... 47 2e-05
TC86873 homologue to GP|530207|gb|AAA66338.1|| heat shock protei... 44 1e-04
TC80686 similar to PIR|T51523|T51523 clpB heat shock protein-lik... 44 1e-04
TC90741 similar to PIR|T51523|T51523 clpB heat shock protein-lik... 41 8e-04
TC93240 weakly similar to GP|14335170|gb|AAK59865.1 AT5g51070/K3... 39 0.003
TC84403 homologue to GP|15021761|gb|AAK77908.1 AAA-metalloprotea... 38 0.009
BG584923 similar to GP|21592785|gb| unknown {Arabidopsis thalian... 35 0.060
TC79768 similar to PIR|B96657|B96657 probable replication factor... 32 0.66
TC81107 similar to SP|O64948|LON1_ARATH Lon protease homolog 1 ... 32 0.66
TC79701 homologue to PIR|T06242|T06242 aspartate kinase (EC 2.7.... 31 0.87
BQ135582 31 0.87
TC93307 similar to GP|9651530|gb|AAF91178.1| ClpB {Phaseolus lun... 31 1.1
>TC76895 homologue to SP|P35100|CLPA_PEA ATP-dependent clp protease
ATP-binding subunit clpA homolog chloroplast precursor.
[Garden pea], complete
Length = 3425
Score = 244 bits (624), Expect(2) = e-110
Identities = 138/304 (45%), Positives = 186/304 (60%)
Frame = +2
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E TE A KVIM AQ+EAR +GH ++ TE ILLG++G+ G+AAK+LKS GI+ +R
Sbjct: 500 MFERFTEKAIKVIMLAQEEARRLGHNFVGTEQILLGLIGEGTGIAAKVLKSMGINLKDAR 679
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
+V K++GRG SGF +I FT AK VL+ SL+ A+ LG++ + + HLLLGLL+
Sbjct: 680 VEVEKIIGRG---SGFVAVEIPFTPRAKRVLELSLEEARQLGHNYIGSEHLLLGLLREGE 850
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
G ++++N GAD IR QVIR + G+ +NVTA +
Sbjct: 851 GVAARVLENLGADPTNIRTQVIRMV-------------------GESADNVTATVGSGS- 970
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
+N + LE +GTNLTKLA+EGKL P VGR+ Q+ERV QI+
Sbjct: 971 -------------------SNNKTPTLEEYGTNLTKLAEEGKLDPVVGRQPQIERVTQIL 1093
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
RR KNNPCL+GEPGVGKT+I +GLAQRI +G VPE ++GKKV+ LD+ + +G
Sbjct: 1094GRRTKNNPCLIGEPGVGKTAIAEGLAQRIANGDVPETIEGKKVITLDMGLLVAGTKYRGE 1273
Query: 301 SEER 304
EER
Sbjct: 1274FEER 1285
Score = 173 bits (438), Expect(2) = e-110
Identities = 90/149 (60%), Positives = 116/149 (77%), Gaps = 1/149 (0%)
Frame = +3
Query: 305 IRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATTVNEH 363
++ L++EI+ ++ILF+ EVH + A A GA A ILK AL RG +QCI ATT++E+
Sbjct: 1287 LKKLMEEIKQSDDIILFIDEVHTLIGAGAAEGAIDAANILKPALARGELQCIGATTLDEY 1466
Query: 364 RMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAANLSQQ 423
R H+E D L+R FQPVKV EP+V+ETI+ILKGLR YE H+KL YTD+AL+AAA LS Q
Sbjct: 1467 RKHIEKDPALERRFQPVKVPEPTVDETIQILKGLRERYEIHHKLRYTDDALIAAAQLSYQ 1646
Query: 424 YVSERFLPDKAIDLIDEAGSHVQLCHAKV 452
Y+S+RFLPDKAIDLIDEAGS V+L HA++
Sbjct: 1647 YISDRFLPDKAIDLIDEAGSRVRLQHAQL 1733
>TC76894 homologue to SP|P35100|CLPA_PEA ATP-dependent clp protease
ATP-binding subunit clpA homolog chloroplast precursor.
[Garden pea], partial (71%)
Length = 2413
Score = 300 bits (769), Expect(2) = e-109
Identities = 171/329 (51%), Positives = 213/329 (63%), Gaps = 1/329 (0%)
Frame = +2
Query: 125 QLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQIKFP 184
++++N GAD IR QVIR + E V T S SN K P
Sbjct: 839 RVLENLGADPTNIRTQVIRMVGEGADSVG-----------------ATVGSGSSNN-KMP 964
Query: 185 VLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQIICRRM 244
L E +GTNLTKLA+EGKL P VGR+ Q+ERV QI+ RR
Sbjct: 965 TL---------------------EEYGTNLTKLAEEGKLDPVVGRQPQIERVTQILGRRT 1081
Query: 245 KNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGSSEER 304
KNNPCL+GEPGVGKT+I +GLAQRI +G VPE ++GKKV+ LD+ + +G EER
Sbjct: 1082 KNNPCLIGEPGVGKTAIAEGLAQRIANGDVPETIEGKKVITLDMGLLVAGTKYRGEFEER 1261
Query: 305 IRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATTVNEH 363
++ L++EI+ +ILF+ EVH + A A GA A ILK AL RG +QCI ATT++E+
Sbjct: 1262 LKKLMEEIKQSDEIILFIDEVHTLIGAGAAEGAIDAANILKPALARGELQCIGATTLDEY 1441
Query: 364 RMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAANLSQQ 423
R H+E D L+R FQPVKV EP+V ETI+ILKGLR YE H+KL YTDEALVAAA LS Q
Sbjct: 1442 RKHIEKDPALERRFQPVKVPEPTVPETIQILKGLRERYEIHHKLRYTDEALVAAAELSHQ 1621
Query: 424 YVSERFLPDKAIDLIDEAGSHVQLCHAKV 452
Y+S+RFLPDKAIDLIDEAGS V+L HA++
Sbjct: 1622 YISDRFLPDKAIDLIDEAGSRVRLQHAQL 1708
Score = 113 bits (282), Expect(2) = e-109
Identities = 60/121 (49%), Positives = 83/121 (68%)
Frame = +1
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E TE A KVIM AQ+EAR +GH ++ TE ILLG++G+ G+AAK+LKS GI+ +R
Sbjct: 475 MFERFTEKAIKVIMLAQEEARRLGHNFVGTEQILLGLIGEGTGIAAKVLKSMGINLKDAR 654
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
+V K++GRG SGF +I FT AK VL+ SL+ A+ LG++ + + H LLGLL+
Sbjct: 655 VEVEKIIGRG---SGFVAVEIPFTPRAKRVLELSLEEARQLGHNYIGSEHSLLGLLREGE 825
Query: 121 G 121
G
Sbjct: 826 G 828
Score = 43.1 bits (100), Expect = 2e-04
Identities = 21/48 (43%), Positives = 31/48 (63%)
Frame = +1
Query: 6 TECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYG 53
T AK+V+ + +EAR +GH YI +EH LLG+L + G+AA +G
Sbjct: 715 TPRAKRVLELSLEEARQLGHNYIGSEHSLLGLLREGEGVAAPCS*KFG 858
Score = 37.0 bits (84), Expect = 0.016
Identities = 22/74 (29%), Positives = 40/74 (53%)
Frame = +1
Query: 68 GRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSNGTVTQLI 127
GRG C + + RFT A V+ + + A+ LG++ V T +LLGL+ G +++
Sbjct: 445 GRGSRCVTKAMFE-RFTEKAIKVIMLAQEEARRLGHNFVGTEQILLGLIGEGTGIAAKVL 621
Query: 128 QNQGADVNKIREQV 141
++ G ++ R +V
Sbjct: 622 KSMGINLKDARVEV 663
>TC81087 homologue to GP|530207|gb|AAA66338.1|| heat shock protein {Glycine
max}, partial (41%)
Length = 1318
Score = 197 bits (501), Expect = 7e-51
Identities = 127/384 (33%), Positives = 204/384 (53%), Gaps = 36/384 (9%)
Frame = +2
Query: 74 SGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSNGTVTQLIQN-QGA 132
S F+ +FT L + + A + G+ + +HL L+ NG Q I N G
Sbjct: 170 SSFTMNPEKFTHKTNEALAGAHELAMTSGHAQITPLHLASILVSDPNGIFFQAISNVAGE 349
Query: 133 DVNKIREQVIRQIEENVHQVNLP----------------AEGSKNQMG-----------G 165
+ + E+V++Q + + + P A+ ++ G G
Sbjct: 350 ESARAVERVLKQALKKLPSQSPPPDEVPGSTALIKAIRRAQAAQKSRGDTHLAVDQLILG 529
Query: 166 QVEENVTAESSKS-----NQIKFPVLTQNNNLGARKDD-NANKQKSALEIFGTNLTKLAQ 219
+E++ + K +++K V G + + + + AL+ +G +L + Q
Sbjct: 530 ILEDSQIGDLFKEAGVAVSRVKTEVEKLRGKDGKKVESASGDTNFQALKTYGRDLVE--Q 703
Query: 220 EGKLHPFVGREEQVERVIQIICRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLK 279
GKL P +GR+E++ RV++I+ RR KNNP L+GEPGVGKT++++GLAQRI+ G VP L
Sbjct: 704 AGKLDPVIGRDEEIRRVVRILSRRTKNNPVLIGEPGVGKTAVVEGLAQRIVRGDVPSNLA 883
Query: 280 GKKVVALDVADFLYVISNQGSSEERIRCLIKEIELC-GNVILFVKEVHHIFDAA-TSGAR 337
+++ALD+ + +G EER++ ++KE+E G VILF+ E+H + A T G+
Sbjct: 884 DVRLIALDMGALVAGAKYRGEFEERLKAVLKEVEEAEGKVILFIDEIHLVLGAGRTEGSM 1063
Query: 338 SFAYILKHALERGVIQCIFATTVNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGL 397
A + K L RG ++CI ATT+ E+R ++E D +R FQ V V EPSV +TI IL+GL
Sbjct: 1064DAANLFKPMLARGQLRCIGATTLEEYRKYVEKDAAFERRFQQVYVAEPSVPDTISILRGL 1243
Query: 398 RGTYESHYKLHYTDEALVAAANLS 421
+ YE H+ + D A+V AA LS
Sbjct: 1244KERYEGHHGVRIQDRAIVVAAQLS 1315
>BG454574 similar to SP|P42762|ERD1 ERD1 protein chloroplast precursor.
[Mouse-ear cress] {Arabidopsis thaliana}, partial (19%)
Length = 672
Score = 140 bits (352), Expect = 1e-33
Identities = 79/190 (41%), Positives = 115/190 (59%), Gaps = 6/190 (3%)
Frame = +1
Query: 194 ARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQIICRRMKNNPCLVGE 253
A K + +K+AL F +LT A G + P +GRE +V+R+IQI+CR+ K+NP L+GE
Sbjct: 103 AAKTKDKKDKKNALSQFCVDLTARASVGLIDPVIGREVEVQRIIQILCRKTKSNPILLGE 282
Query: 254 PGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGSSEERIRCLIKEIE 313
GVGKT+I +GLA I V L K+V++LDV + +G E+R+ LIK+I
Sbjct: 283 AGVGKTAIAEGLAILISRAEVAPFLLTKRVMSLDVGLLMAGAKERGELEDRVTKLIKDII 462
Query: 314 LCGNVILFVKEVHHIFDAATSGARS------FAYILKHALERGVIQCIFATTVNEHRMHM 367
G+VILF+ EVH + + T+G + A +LK +L RG QCI +TT++E+R+H
Sbjct: 463 ESGDVILFIDEVHTLVQSGTTGRGNKGSGFDIANLLKPSLGRGQFQCIASTTIDEYRLHF 642
Query: 368 ENDTTLKRIF 377
E D L F
Sbjct: 643 EKDKALAXRF 672
>TC85056 homologue to GP|9651530|gb|AAF91178.1| ClpB {Phaseolus lunatus},
partial (25%)
Length = 743
Score = 133 bits (335), Expect = 1e-31
Identities = 68/133 (51%), Positives = 96/133 (72%), Gaps = 1/133 (0%)
Frame = +1
Query: 316 GNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATTVNEHRMHMENDTTLK 374
G +ILF+ E+H + A ATSGA +LK L RG ++CI ATT+NE+R ++E D L+
Sbjct: 16 GQIILFIDEIHTVVGAGATSGAMDAGNLLKPMLGRGELRCIGATTLNEYRKYIEKDPALE 195
Query: 375 RIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAANLSQQYVSERFLPDKA 434
R FQ V +PSVE+TI IL+GLR YE H+ + +D ALV+AA L+ +Y++ERFLPDKA
Sbjct: 196 RRFQQVFCCQPSVEDTISILRGLRERYELHHGVKISDSALVSAAVLADRYITERFLPDKA 375
Query: 435 IDLIDEAGSHVQL 447
IDL+DEA + +++
Sbjct: 376 IDLVDEAAAKLKM 414
>TC76897 homologue to SP|P35100|CLPA_PEA ATP-dependent clp protease
ATP-binding subunit clpA homolog chloroplast precursor.
[Garden pea], partial (18%)
Length = 826
Score = 128 bits (321), Expect = 5e-30
Identities = 69/147 (46%), Positives = 95/147 (63%)
Frame = +2
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E TE A KVIM AQ+EAR +GH ++ TE ILLG++G+ G+ AK+LKS GI+ SR
Sbjct: 374 MFERFTEKAIKVIMLAQEEARRLGHNFVGTEQILLGLIGEGTGIGAKVLKSMGINLKDSR 553
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
+V K++GRG SGF +I FT AK VL+ SL+ A+ LG++ + + HLLLGLL+
Sbjct: 554 VEVEKIIGRG---SGFVAVEIPFTPRAKRVLELSLEEARQLGHNYIGSEHLLLGLLREGE 724
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEE 147
G ++++N GAD I VI E
Sbjct: 725 GVAARVLENLGADPTNIXTXVIXMXGE 805
Score = 55.1 bits (131), Expect = 6e-08
Identities = 27/70 (38%), Positives = 43/70 (60%)
Frame = +2
Query: 6 TECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSREQVMK 65
T AK+V+ + +EAR +GH YI +EH+LLG+L + G+AA++L++ G D V+
Sbjct: 614 TPRAKRVLELSLEEARQLGHNYIGSEHLLLGLLREGEGVAARVLENLGADPTNIXTXVIX 793
Query: 66 LVGRGGGCSG 75
+ G G G
Sbjct: 794 MXGEGADNXG 823
>BQ752143 similar to PIR|T39572|T39 probable proteinase subunit - fission
yeast (Schizosaccharomyces pombe), partial (18%)
Length = 774
Score = 112 bits (281), Expect = 2e-25
Identities = 56/130 (43%), Positives = 90/130 (69%), Gaps = 1/130 (0%)
Frame = +2
Query: 196 KDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQIICRRMKNNPCLVGEPG 255
K+ + ++ L F ++T LA++ K+ P +GREE++ RV++I+ RR KNNP L+GEPG
Sbjct: 278 KNADTEEEHENLAKFTIDMTALARDKKIDPVIGREEEIRRVVRILSRRTKNNPVLIGEPG 457
Query: 256 VGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGSSEERIRCLIKEIELC 315
VGKT++++GLAQRI++ VP+ LK K+++LDV + +G EER++ ++KEIE
Sbjct: 458 VGKTTVVEGLAQRIVNRDVPDNLKACKLLSLDVGALVAGSKYRGEFEERMKGVLKEIEDS 637
Query: 316 GN-VILFVKE 324
++LFV E
Sbjct: 638 KEMIVLFVDE 667
>TC90378 weakly similar to GP|21592785|gb|AAM64734.1 unknown {Arabidopsis
thaliana}, partial (45%)
Length = 950
Score = 49.7 bits (117), Expect = 2e-06
Identities = 35/131 (26%), Positives = 61/131 (45%)
Frame = +1
Query: 9 AKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSREQVMKLVG 68
A K ++ E R + + + TE +L+GIL + AAK L++ GI + REQ ++++G
Sbjct: 322 AIKAYAMSELETRKLKYPKLGTEALLMGILIEGTSKAAKFLRANGITYLKVREQTLEILG 501
Query: 69 RGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSNGTVTQLIQ 128
+ S S + T LD++L G + HLLLG+ +++
Sbjct: 502 K-SYWSYSSPVVLSLTEPLHKALDWALNETSKSGEGETNVTHLLLGIWSQEESAGRKVLS 678
Query: 129 NQGADVNKIRE 139
G + K +E
Sbjct: 679 ALGFNDQKAKE 711
>TC90938 similar to GP|21592785|gb|AAM64734.1 unknown {Arabidopsis
thaliana}, partial (53%)
Length = 691
Score = 46.6 bits (109), Expect = 2e-05
Identities = 34/124 (27%), Positives = 64/124 (51%), Gaps = 3/124 (2%)
Frame = +2
Query: 19 EARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSREQVMKLVGRGGGCSGFSC 78
EAR + TE +++GIL + LA+K L++ GI R++++K++G F+
Sbjct: 326 EARKLKFTTTGTEALIMGILIEGTNLASKFLRANGITLFKVRDEIVKILGE---VDPFTE 496
Query: 79 KDIR--FTFDAKNVLDFSL-KHAKSLGYDNVDTMHLLLGLLQGSNGTVTQLIQNQGADVN 135
R T DA+ LD+++ + KS+ V T H++LG+ + +++ G +
Sbjct: 497 TPERPPMTDDAQRALDWAVDRKLKSVDGGEVTTAHIILGIWSDVDSPGHKILSKLGINDE 676
Query: 136 KIRE 139
K +E
Sbjct: 677 KAKE 688
Score = 29.6 bits (65), Expect = 2.5
Identities = 17/58 (29%), Positives = 32/58 (54%), Gaps = 1/58 (1%)
Frame = +2
Query: 5 LTECAKKVI-MAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSRE 61
+T+ A++ + A ++ + V ++T HI+LGI D + KIL GI+ ++E
Sbjct: 515 MTDDAQRALDWAVDRKLKSVDGGEVTTAHIILGIWSDVDSPGHKILSKLGINDEKAKE 688
>TC86873 homologue to GP|530207|gb|AAA66338.1|| heat shock protein {Glycine
max}, partial (53%)
Length = 1475
Score = 44.3 bits (103), Expect = 1e-04
Identities = 20/35 (57%), Positives = 29/35 (82%)
Frame = +2
Query: 413 ALVAAANLSQQYVSERFLPDKAIDLIDEAGSHVQL 447
A+V AA LS +Y++ R LPDKAIDL+DEA ++V++
Sbjct: 2 AIVVAAQLSSRYITGRHLPDKAIDLVDEACANVRV 106
>TC80686 similar to PIR|T51523|T51523 clpB heat shock protein-like -
Arabidopsis thaliana, partial (21%)
Length = 982
Score = 44.3 bits (103), Expect = 1e-04
Identities = 31/115 (26%), Positives = 53/115 (45%)
Frame = +1
Query: 3 ENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSREQ 62
+ TE A + I+++ + A+ H+ + TEH++ +L NGLA +I G+D E
Sbjct: 343 QEFTEMAWQAIVSSPEVAKENKHQIVETEHLMKALLEQKNGLARRIFTKVGVDNTRLLEA 522
Query: 63 VMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQ 117
K + R G S + D + ++ + + K G V HL+LG Q
Sbjct: 523 TDKHIQRQPKVLGESAGSM-LGRDLEALIQRAREFKKEYGDSFVSVEHLVLGFAQ 684
Score = 42.7 bits (99), Expect = 3e-04
Identities = 18/46 (39%), Positives = 31/46 (67%)
Frame = +2
Query: 197 DDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQIICR 242
D + + ALE +G +LT +A+ GKL P +GR++++ R IQI+ +
Sbjct: 785 DQDPEGKYEALEKYGKDLTAMAKAGKLDPVIGRDDEIRRCIQILSK 922
Score = 42.0 bits (97), Expect = 5e-04
Identities = 57/255 (22%), Positives = 97/255 (37%), Gaps = 35/255 (13%)
Frame = +1
Query: 42 NGLAAKILKSYGIDFNVSREQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSL 101
N + K +++ F+ R + + R SG FT A + S + AK
Sbjct: 229 NSIPLKKREAFSNGFSRRRRNSKQFIVRCTDSSG-KVSQQEFTEMAWQAIVSSPEVAKEN 405
Query: 102 GYDNVDTMHLLLGLLQGSNGTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKN 161
+ V+T HL+ LL+ NG ++ G D ++ E + I+ Q + E + +
Sbjct: 406 KHQIVETEHLMKALLEQKNGLARRIFTKVGVDNTRLLEATDKHIQR---QPKVLGESAGS 576
Query: 162 QMGGQVEENVT-AESSKSNQIKFPVLTQNNNLGARKDDNANKQ------------KSALE 208
+G +E + A K V ++ LG +D K K+
Sbjct: 577 MLGRDLEALIQRAREFKKEYGDSFVSVEHLVLGFAQDRRFGKDIV*RFSNISTSSKNCYR 756
Query: 209 IF-GTNLT-------KLAQEGKLHPFVGREEQVERVIQIICRR--------------MKN 246
+ GT ++ K+ GK+ Q ++ +R KN
Sbjct: 757 VRKGTPISY*SRS*RKIRSLGKIRKRFDSNGQSRKIRSCYRKR**NTKVHSNSLQGRTKN 936
Query: 247 NPCLVGEPGVGKTSI 261
NP L+GEPGVGKT++
Sbjct: 937 NPLLIGEPGVGKTAL 981
>TC90741 similar to PIR|T51523|T51523 clpB heat shock protein-like -
Arabidopsis thaliana, partial (24%)
Length = 710
Score = 41.2 bits (95), Expect = 8e-04
Identities = 18/30 (60%), Positives = 25/30 (83%)
Frame = +1
Query: 418 ANLSQQYVSERFLPDKAIDLIDEAGSHVQL 447
A LS +Y+S RFLPDKAIDL+DEA + +++
Sbjct: 52 AILSDRYISGRFLPDKAIDLVDEAAAKLKM 141
>TC93240 weakly similar to GP|14335170|gb|AAK59865.1 AT5g51070/K3K7_27
{Arabidopsis thaliana}, partial (8%)
Length = 700
Score = 39.3 bits (90), Expect = 0.003
Identities = 31/129 (24%), Positives = 58/129 (44%), Gaps = 16/129 (12%)
Frame = +1
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILG---DSNGLAAKI--------- 48
+ E TE + K I+ A+KEA+F ++ +HI+LG++ +SN L+ +
Sbjct: 325 IFERFTERSIKSIVYAEKEAKFFKSDFLYAQHIMLGLIAEAEESNRLSKRFS*FWCSHS* 504
Query: 49 ----LKSYGIDFNVSREQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYD 104
S FN R + C F+F K V + ++++++SL ++
Sbjct: 505 ESS*CCSSSQ*FN*LRLMITV-------CMSIKIDPFPFSFGTKRVFEAAVEYSRSLNHN 663
Query: 105 NVDTMHLLL 113
VD H+ +
Sbjct: 664 FVDPEHIFV 690
>TC84403 homologue to GP|15021761|gb|AAK77908.1 AAA-metalloprotease FtsH
{Pisum sativum}, partial (36%)
Length = 897
Score = 37.7 bits (86), Expect = 0.009
Identities = 26/79 (32%), Positives = 37/79 (45%)
Frame = +3
Query: 250 LVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGSSEERIRCLI 309
LVG PG GKT + + A G +++ +DFL + GSS R+R L
Sbjct: 30 LVGPPGTGKTLLAKATAGE----------SGVPFLSISGSDFLELFVGVGSS--RVRNLF 173
Query: 310 KEIELCGNVILFVKEVHHI 328
KE C I+F+ E+ I
Sbjct: 174 KEARKCAPSIVFIDEIDAI 230
>BG584923 similar to GP|21592785|gb| unknown {Arabidopsis thaliana}, partial
(27%)
Length = 587
Score = 35.0 bits (79), Expect = 0.060
Identities = 17/50 (34%), Positives = 31/50 (62%)
Frame = +1
Query: 19 EARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSREQVMKLVG 68
EAR + TE +++GIL + LA+K L++ GI R++++K++G
Sbjct: 229 EARKLKFTTTGTEALIMGILIEGTNLASKFLRANGITLFKVRDEIVKILG 378
>TC79768 similar to PIR|B96657|B96657 probable replication factor F16M19.6
[imported] - Arabidopsis thaliana, complete
Length = 1246
Score = 31.6 bits (70), Expect = 0.66
Identities = 23/57 (40%), Positives = 31/57 (54%), Gaps = 2/57 (3%)
Frame = +1
Query: 222 KLHPFVGREEQVERVIQIICRRMKNNPCLV--GEPGVGKTSIIQGLAQRILSGSVPE 276
K+ VG E+ V R +Q+I R N P L+ G PG GKT+ I LA +L + E
Sbjct: 154 KVVDIVGNEDAVSR-LQVIARD-GNMPNLILSGPPGTGKTTSILALAHELLGPNYRE 318
>TC81107 similar to SP|O64948|LON1_ARATH Lon protease homolog 1
mitochondrial precursor (EC 3.4.21.-). [Mouse-ear
cress], partial (45%)
Length = 1266
Score = 31.6 bits (70), Expect = 0.66
Identities = 25/73 (34%), Positives = 36/73 (49%), Gaps = 1/73 (1%)
Frame = +2
Query: 249 CLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFL-YVISNQGSSEERIRC 307
C VG PGVGKTS+ +A + V L G K D AD + + GS R+
Sbjct: 584 CFVGPPGVGKTSLASSIAAALDRKFVRISLGGVK----DEADIRGHRRTYIGSMPGRLID 751
Query: 308 LIKEIELCGNVIL 320
+K++ +C V+L
Sbjct: 752 GLKKVAVCNPVML 790
>TC79701 homologue to PIR|T06242|T06242 aspartate kinase (EC 2.7.2.4) /
homoserine dehydrogenase (EC 1.1.1.3) precursor -
soybean, partial (28%)
Length = 1042
Score = 31.2 bits (69), Expect = 0.87
Identities = 23/84 (27%), Positives = 34/84 (40%), Gaps = 7/84 (8%)
Frame = +1
Query: 194 ARKDDNANKQKSALEIFGT-------NLTKLAQEGKLHPFVGREEQVERVIQIICRRMKN 246
A+K ++A+ L G + +L + K HPF + +I RR KN
Sbjct: 499 AKKQEDADNAGEVLRYVGVVDVTNKKGVVELRKYKKDHPFAQLSGS-DNIIAFTTRRYKN 675
Query: 247 NPCLVGEPGVGKTSIIQGLAQRIL 270
P +V PG G G+ IL
Sbjct: 676 QPLIVRGPGAGAQVTAGGIFSDIL 747
>BQ135582
Length = 733
Score = 31.2 bits (69), Expect = 0.87
Identities = 22/67 (32%), Positives = 28/67 (40%), Gaps = 3/67 (4%)
Frame = -2
Query: 315 CGNVILFVKEVHHI---FDAATSGARSFAYILKHALERGVIQCIFATTVNEHRMHMENDT 371
C N +LF K+V H F R + L L V FA EHR M+N
Sbjct: 558 CSNRVLF*KQVVHASGQFSPVRIRVRKYRSFLVVRLSSSVPVFGFACDKREHRSVMDNQQ 379
Query: 372 TLKRIFQ 378
+L +FQ
Sbjct: 378 SLNPLFQ 358
>TC93307 similar to GP|9651530|gb|AAF91178.1| ClpB {Phaseolus lunatus},
partial (11%)
Length = 607
Score = 30.8 bits (68), Expect = 1.1
Identities = 15/50 (30%), Positives = 27/50 (54%)
Frame = +2
Query: 6 TECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGID 55
TE A + ++ A AR + + +EH++ +L +GLA +I G+D
Sbjct: 449 TEMAWEGVIGAVDAARVNKQQIVESEHLMKALLEQRDGLARRIFTKAGLD 598
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.135 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,701,520
Number of Sequences: 36976
Number of extensions: 142029
Number of successful extensions: 763
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 735
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 750
length of query: 455
length of database: 9,014,727
effective HSP length: 99
effective length of query: 356
effective length of database: 5,354,103
effective search space: 1906060668
effective search space used: 1906060668
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146751.5


