

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146751.12 - phase: 0
(513 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
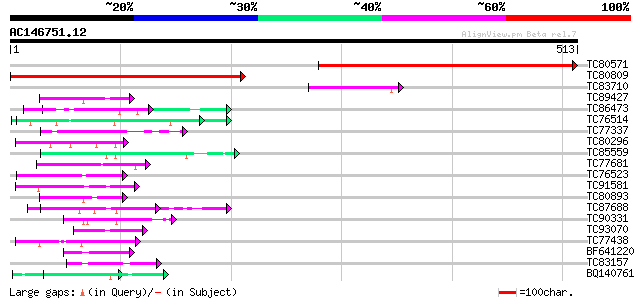
Score E
Sequences producing significant alignments: (bits) Value
TC80571 similar to GP|22136852|gb|AAM91770.1 unknown protein {Ar... 464 e-131
TC80809 similar to GP|22136852|gb|AAM91770.1 unknown protein {Ar... 422 e-118
TC83710 similar to PIR|A86201|A86201 protein F12K11.11 [imported... 56 4e-08
TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed pr... 54 1e-07
TC86473 similar to PIR|E84828|E84828 probable WD-40 repeat prote... 51 9e-07
TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 49 6e-06
TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Atel... 46 3e-05
TC80296 similar to PIR|H86265|H86265 protein F3F19.18 [imported]... 46 4e-05
TC85559 similar to PIR|T07012|T07012 acetyl-CoA carboxylase (EC ... 45 5e-05
TC77681 similar to GP|22597156|gb|AAN03465.1 nucleolar histone d... 45 9e-05
TC76523 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 44 1e-04
TC91581 weakly similar to GP|7269000|emb|CAB80733.1 contains EST... 44 2e-04
TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {... 44 2e-04
TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~un... 44 2e-04
TC90331 weakly similar to GP|21751020|dbj|BAC03887. unnamed prot... 44 2e-04
TC93070 weakly similar to GP|17065230|gb|AAL32769.1 Unknown prot... 43 3e-04
TC77438 homologue to PIR|T06807|T06807 nucleosome assembly prote... 43 3e-04
BF641220 weakly similar to PIR|G86203|G86 probable N-arginine di... 43 3e-04
TC83157 similar to GP|21595368|gb|AAM66095.1 unknown {Arabidopsi... 43 3e-04
BQ140761 weakly similar to GP|18076245|emb phosphophoryn {Rattus... 42 4e-04
>TC80571 similar to GP|22136852|gb|AAM91770.1 unknown protein {Arabidopsis
thaliana}, partial (35%)
Length = 1502
Score = 464 bits (1193), Expect = e-131
Identities = 232/234 (99%), Positives = 232/234 (99%)
Frame = +1
Query: 280 VPIGATPEYMAGFYMLQSASSFLPVMALAPQEKERIVDMAAAPGGKTTYIAALMKNSGII 339
VPIGATPEYMAGFYMLQSASSFLPVMALAPQEKERIVDMAAAPGGKTTYIAALMKNSGII
Sbjct: 1 VPIGATPEYMAGFYMLQSASSFLPVMALAPQEKERIVDMAAAPGGKTTYIAALMKNSGII 180
Query: 340 FANEMKVPRLKSLTANLHRMGVSNTVVSNYDGKELPKVLGFNSVDRVLLDAPCSGTGVIS 399
FANEMKVPRLKSLTANLHRMGVSNTVVSNYDGKELPKVLGFNSVDRVLLDAPCSGTGVIS
Sbjct: 181 FANEMKVPRLKSLTANLHRMGVSNTVVSNYDGKELPKVLGFNSVDRVLLDAPCSGTGVIS 360
Query: 400 KDESVKTSKNLEDIKKCAHLQKELLLAAIDMVDSYSKSGGYVVYSTCSIMVAENEAVIDY 459
KDESVKTSKNLEDIKKCAHLQKELLLAAIDMVDSYSKSGGYVVYSTCSIMVAENEAVIDY
Sbjct: 361 KDESVKTSKNLEDIKKCAHLQKELLLAAIDMVDSYSKSGGYVVYSTCSIMVAENEAVIDY 540
Query: 460 VLKRRDVKLVPCGLDFGRPGFTKFREQRFHPSLDKTRRFYPHVHNMDGFFVAKV 513
VLKRRDVKLVPCGLDFGRPGFTKFREQRFHPSLDKTRRFYPHVHNMDGFF KV
Sbjct: 541 VLKRRDVKLVPCGLDFGRPGFTKFREQRFHPSLDKTRRFYPHVHNMDGFFCCKV 702
>TC80809 similar to GP|22136852|gb|AAM91770.1 unknown protein {Arabidopsis
thaliana}, partial (11%)
Length = 681
Score = 422 bits (1085), Expect = e-118
Identities = 213/213 (100%), Positives = 213/213 (100%)
Frame = +3
Query: 1 MAPATKKKPSKPVKPQKAPKKVQIVPIKDDSTSESEEEEAPPLQHLELDGSDSDLSSDGD 60
MAPATKKKPSKPVKPQKAPKKVQIVPIKDDSTSESEEEEAPPLQHLELDGSDSDLSSDGD
Sbjct: 42 MAPATKKKPSKPVKPQKAPKKVQIVPIKDDSTSESEEEEAPPLQHLELDGSDSDLSSDGD 221
Query: 61 DQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEEDIARKSKLIDRKRE 120
DQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEEDIARKSKLIDRKRE
Sbjct: 222 DQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEEDIARKSKLIDRKRE 401
Query: 121 RDSQAAADELQTNIQTNIQDESDEFTLPTKQELEEEALRPPDLSNLQRRIKEIVRVLSNF 180
RDSQAAADELQTNIQTNIQDESDEFTLPTKQELEEEALRPPDLSNLQRRIKEIVRVLSNF
Sbjct: 402 RDSQAAADELQTNIQTNIQDESDEFTLPTKQELEEEALRPPDLSNLQRRIKEIVRVLSNF 581
Query: 181 KALRQDGATRKDYVDQLKTDIRSYYGYNEFLIG 213
KALRQDGATRKDYVDQLKTDIRSYYGYNEFLIG
Sbjct: 582 KALRQDGATRKDYVDQLKTDIRSYYGYNEFLIG 680
>TC83710 similar to PIR|A86201|A86201 protein F12K11.11 [imported] -
Arabidopsis thaliana, partial (39%)
Length = 934
Score = 55.8 bits (133), Expect = 4e-08
Identities = 35/89 (39%), Positives = 49/89 (54%), Gaps = 3/89 (3%)
Frame = +1
Query: 271 VGLVVYDSQVPIGATPEYMAGFYMLQSASSFLPVMALAPQEKERIVDMAAAPGGKTTYIA 330
+G+ + D + + + G LQ+ S + AL PQ ERI+DM AAPGGKTT IA
Sbjct: 286 LGVDMKDRVYELHSFHNVLEGEIFLQNLPSIIAAHALDPQMGERILDMCAAPGGKTTAIA 465
Query: 331 ALMKNSGIIFANEM---KVPRLKSLTANL 356
LMK+ G + A + KV ++ L A L
Sbjct: 466 ILMKDKGEVIATDRSHNKVLDIEKLAAEL 552
Score = 28.9 bits (63), Expect = 5.0
Identities = 12/17 (70%), Positives = 14/17 (81%)
Frame = +1
Query: 381 NSVDRVLLDAPCSGTGV 397
+S DRVLLDAPCS G+
Sbjct: 877 DSFDRVLLDAPCSALGL 927
>TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed protein
{Arabidopsis thaliana}, partial (50%)
Length = 753
Score = 54.3 bits (129), Expect = 1e-07
Identities = 34/94 (36%), Positives = 50/94 (53%), Gaps = 8/94 (8%)
Frame = +1
Query: 28 KDDSTSESEEEEAPPLQ-HLELDGSDSDLSSDGDDQLAD-------DFLQGSDDEDNDND 79
KD S +E +++E + E D +D D S D DD+ AD + GSDD+D D+D
Sbjct: 292 KDGSETEDDDDEDDDDDVNDEDDDNDEDFSGDEDDEDADPEDDPVPNGAGGSDDDDEDDD 471
Query: 80 NDDEGSDLESGSGSDEDEDSDSESGEEDIARKSK 113
+DD+ +D G DEDED + + E+ K K
Sbjct: 472 DDDDDND----DGEDEDEDEEEDDDEDQPPSKKK 561
Score = 40.0 bits (92), Expect = 0.002
Identities = 25/82 (30%), Positives = 40/82 (48%), Gaps = 8/82 (9%)
Frame = +1
Query: 49 DGSDSDLSSDGDDQLADDFLQGSDDEDND--NDNDDEGSDLES------GSGSDEDEDSD 100
DGS+++ D DD DD DD D D D DDE +D E GSD+D++ D
Sbjct: 295 DGSETEDDDDEDDD--DDVNDEDDDNDEDFSGDEDDEDADPEDDPVPNGAGGSDDDDEDD 468
Query: 101 SESGEEDIARKSKLIDRKRERD 122
+ +++ + + D + + D
Sbjct: 469 DDDDDDNDDGEDEDEDEEEDDD 534
Score = 36.6 bits (83), Expect = 0.024
Identities = 28/111 (25%), Positives = 49/111 (43%)
Frame = +1
Query: 42 PLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDS 101
PL L + + DG + DD DD+D ++++DD D SG ++DED+D
Sbjct: 247 PLDQLSKGNHTCEENKDGSETEDDD--DEDDDDDVNDEDDDNDEDF---SGDEDDEDADP 411
Query: 102 ESGEEDIARKSKLIDRKRERDSQAAADELQTNIQTNIQDESDEFTLPTKQE 152
E D + + D D+ + + +D+ DE P+K++
Sbjct: 412 EDDPVPNGAGGSDDDDEDDDDDDDDNDDGEDEDEDEEEDD-DEDQPPSKKK 561
>TC86473 similar to PIR|E84828|E84828 probable WD-40 repeat protein
[imported] - Arabidopsis thaliana, partial (68%)
Length = 2654
Score = 51.2 bits (121), Expect = 9e-07
Identities = 34/118 (28%), Positives = 53/118 (44%)
Frame = +2
Query: 13 VKPQKAPKKVQIVPIKDDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSD 72
+ P KK +V + +E+EEAPP SD D D DD +DD ++ SD
Sbjct: 17 LSPSSTMKKKTVVESEMKKKKSNEKEEAPP------PPSDHD---DSDDDDSDDSVEVSD 169
Query: 73 DEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEEDIARKSKLIDRKRERDSQAAADEL 130
+ D D ++ SD D DS SE +E+ A + + + DSQ +++
Sbjct: 170 YDSISEDEDSSADSIDDDDSSDSDSDSKSELEDEEGASPTTHHNASDDSDSQDEEEDV 343
Score = 44.3 bits (103), Expect = 1e-04
Identities = 44/193 (22%), Positives = 77/193 (39%), Gaps = 22/193 (11%)
Frame = +2
Query: 30 DSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLES 89
DS SE E+ A + + SDSD S+ +D+ + D D+D+ DE D+
Sbjct: 173 DSISEDEDSSADSIDDDDSSDSDSDSKSELEDEEGASPTTHHNASD-DSDSQDEEEDVVD 349
Query: 90 GSGSDEDED-------SDSESGEEDIARKSKL---------------IDRKRERDSQAAA 127
G E D ++S+S E+++A ++ + D K ++ +
Sbjct: 350 SEGGSESSDLHQEGGGAESDSSEDEVAPRNTIGEVPLKWYEDEPHIGYDIKGKKIKKKER 529
Query: 128 DELQTNIQTNIQDESDEFTLPTKQELEEEALRPPDLSNLQRRIKEIVRVLSNFKALRQDG 187
++ + N+ D + + + EE L ++ ++VR L KA D
Sbjct: 530 EDKLGSFLANVDDSKNWRKVFDEYNDEEVVLTKDEI--------KMVRKLMQNKAPHSDF 685
Query: 188 ATRKDYVDQLKTD 200
DYVD K D
Sbjct: 686 DPHPDYVDWFKWD 724
>TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (61%)
Length = 1288
Score = 48.5 bits (114), Expect = 6e-06
Identities = 46/188 (24%), Positives = 75/188 (39%), Gaps = 18/188 (9%)
Frame = +3
Query: 7 KKPSKPVKPQK-----APKKVQIVPIKDDSTSESEEEEAP------------PLQHLELD 49
KKP+ P K A K D+S+ E EE+E P P + + +
Sbjct: 564 KKPAAKAVPSKNGSAPAKKAASDEEDTDESSDEDEEDEKPAAKAVPSKNGSVPAKKADTE 743
Query: 50 GSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEEDIA 109
SD D S D++ + S + SD ES SDEDED+ S +A
Sbjct: 744 SSDED-SESSDEEDKKPAAKASKNVSAPTKKAASSSDEESDEESDEDEDAKPVSKPAAVA 920
Query: 110 RKSKL-IDRKRERDSQAAADELQTNIQTNIQDESDEFTLPTKQELEEEALRPPDLSNLQR 168
+KSK + D +++DE + + +D+ + + + EE Q+
Sbjct: 921 KKSKKDSSDSDDEDDDSSSDEDKKPVAAKKEDKMNVDKDGSDSDQSEEESEDEPSKTPQK 1100
Query: 169 RIKEIVRV 176
+IK++ V
Sbjct: 1101KIKDVEMV 1124
Score = 44.7 bits (104), Expect = 9e-05
Identities = 49/218 (22%), Positives = 81/218 (36%), Gaps = 19/218 (8%)
Frame = +3
Query: 2 APA-TKKKPSKPVKPQKAPKKVQIVPIKDDSTSESEEEEAPPLQHLELDGSDSDLSSDGD 60
APA +KK P+K +KA + DS+S EEE P+ + S + D
Sbjct: 342 APAPSKKTPAKKGNVKKAQPETTSEESDSDSSSSDEEEVKKPVSKAVPSKNGSAPAKKVD 521
Query: 61 DQLADDFLQGSDDEDNDNDNDDEGSDLESGSGS---------DEDEDSDSESGEEDIARK 111
+D +E +D D + S +GS +ED D S+ EED
Sbjct: 522 TSEEED-----SEESSDEDKKPAAKAVPSKNGSAPAKKAASDEEDTDESSDEDEEDEKPA 686
Query: 112 SKLIDRKRERDSQAAADELQTNIQTNIQDESDE---------FTLPTKQELEEEALRPPD 162
+K + K AD ++ + DE D+ + PTK+ +
Sbjct: 687 AKAVPSKNGSVPAKKADTESSDEDSESSDEEDKKPAAKASKNVSAPTKKAASSSDEESDE 866
Query: 163 LSNLQRRIKEIVRVLSNFKALRQDGATRKDYVDQLKTD 200
S+ K + + + K ++D + D D +D
Sbjct: 867 ESDEDEDAKPVSKPAAVAKKSKKDSSDSDDEDDDSSSD 980
Score = 32.0 bits (71), Expect = 0.59
Identities = 32/123 (26%), Positives = 51/123 (41%), Gaps = 22/123 (17%)
Frame = +3
Query: 6 KKKPSKPVKPQKAPKKVQIVPIKDDSTSESEEEE-APPLQH------------LELDGSD 52
KK +K K AP K ++S ES+E+E A P+ + D D
Sbjct: 783 KKPAAKASKNVSAPTKKAASSSDEESDEESDEDEDAKPVSKPAAVAKKSKKDSSDSDDED 962
Query: 53 SDLSSDGD---------DQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSES 103
D SSD D D++ D GSD + ++ +++DE S D + +S
Sbjct: 963 DDSSSDEDKKPVAAKKEDKMNVD-KDGSDSDQSEEESEDEPSKTPQKKIKDVEMVDAGKS 1139
Query: 104 GEE 106
G++
Sbjct: 1140GKK 1148
>TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Ateline
herpesvirus 3}, partial (8%)
Length = 986
Score = 46.2 bits (108), Expect = 3e-05
Identities = 34/133 (25%), Positives = 59/133 (43%)
Frame = +3
Query: 29 DDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLE 88
D + E E+E L + G ++ ++ + + A + G DE+ + ++D++G D +
Sbjct: 462 DGAFGEGEDE----LSSEDGGGYGNNSNNKSNSKKAPEGGAGGADENGEEEDDEDGDDQD 629
Query: 89 SGSGSDEDEDSDSESGEEDIARKSKLIDRKRERDSQAAADELQTNIQTNIQDESDEFTLP 148
DED+D + E GEE D + DE + N ++E DE
Sbjct: 630 EDDDEDEDDDDEEEGGEE---------------DEEEGVDE-----EDNEEEEEDE---- 737
Query: 149 TKQELEEEALRPP 161
+EEAL+PP
Sbjct: 738 -----DEEALQPP 761
Score = 35.8 bits (81), Expect = 0.041
Identities = 29/117 (24%), Positives = 45/117 (37%)
Frame = +3
Query: 40 APPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDS 99
AP L L L +D + + D ++ ED ++D D+E D + G EDE S
Sbjct: 324 APELYALML--TDMPTTKGDSKTQSQDKQHDAEGEDGNDDEDEEDDDGDGAFGEGEDELS 497
Query: 100 DSESGEEDIARKSKLIDRKRERDSQAAADELQTNIQTNIQDESDEFTLPTKQELEEE 156
+ G +K +K ADE D+ DE + + +EE
Sbjct: 498 SEDGGGYGNNSNNKSNSKKAPEGGAGGADENGEEEDDEDGDDQDEDDDEDEDDDDEE 668
>TC80296 similar to PIR|H86265|H86265 protein F3F19.18 [imported] -
Arabidopsis thaliana, partial (34%)
Length = 1734
Score = 45.8 bits (107), Expect = 4e-05
Identities = 39/122 (31%), Positives = 52/122 (41%), Gaps = 20/122 (16%)
Frame = +3
Query: 6 KKKPSKPVKPQKAPKKVQIVPIKDDSTSES-----EEEEAPPLQHLELDGSDS------- 53
KK +P P+ PK V + D +++ H + GSD+
Sbjct: 651 KKDRGRPTDPKAKPKAYGEVNVAADVPGAELLQIIDDDVEQESSHSDDCGSDNAQEDDQV 830
Query: 54 DLSSDGDDQLADDFLQGSDDEDND-----NDNDDEGSDLE-SGSGSD--EDEDSDSESGE 105
L+SD D+QL D DDE D +D +D SD E SG +D EDE D E E
Sbjct: 831 SLNSDDDNQLGSDNTGSDDDEAEDHDGVSDDENDRSSDYETSGDDADNVEDEGDDLEDSE 1010
Query: 106 ED 107
ED
Sbjct: 1011ED 1016
>TC85559 similar to PIR|T07012|T07012 acetyl-CoA carboxylase (EC 6.4.1.2) -
potato chloroplast, partial (66%)
Length = 5011
Score = 45.4 bits (106), Expect = 5e-05
Identities = 48/203 (23%), Positives = 80/203 (38%), Gaps = 23/203 (11%)
Frame = +3
Query: 29 DDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLAD-DFLQ-GSDDEDNDNDNDDEGSD 86
DD + + P+ + + SDSD SD +D D D LQ G+D + +NDND + D
Sbjct: 2736 DDDKLQKASDILEPVNYSAENYSDSDNDSDDNDPDDDYDTLQKGTDTLEPENDNDTDDYD 2915
Query: 87 --------LESGSGSD-----------EDEDSDSESGEEDIARKSKLIDRKRERDSQAAA 127
LE + SD +DE D ++ + + S +++ + DS+
Sbjct: 2916 KIQRAHYILEPENDSDTEPDDEPDYEPDDEPDDEPDDDDKLQKASDILEPENPSDSEKVQ 3095
Query: 128 DELQTNIQTNIQDESDEFTLPTKQELEEEAL--RPPDLSNLQRRIKEIVRVLSNFKALRQ 185
+ + +++ SD + E+E + P D +Q N +
Sbjct: 3096 NSSEIESTGIMENPSDSEKVQNSSEIESTGIMENPSDSEKVQ-----------NSSEIES 3242
Query: 186 DGATRKDYVDQLKTDIRSYYGYN 208
G RKD+ L S YG N
Sbjct: 3243 TGIIRKDF-SHLWVACDSCYGNN 3308
Score = 38.9 bits (89), Expect = 0.005
Identities = 35/114 (30%), Positives = 53/114 (45%), Gaps = 6/114 (5%)
Frame = +3
Query: 29 DDSTSES--EEEEAPPLQHLELDGSDSDLSSD---GDDQLADDFLQ-GSDDEDNDNDNDD 82
+D SE E+E+ P LE + S++ L + ++ L D+ + G +DED ++
Sbjct: 3468 EDEPSEKGLEDEDEPSENSLEDEPSENSLEDEDEPSENSLEDEPSEKGLEDEDEPSEKGL 3647
Query: 83 EGSDLESGSGSDEDEDSDSESGEEDIARKSKLIDRKRERDSQAAADELQTNIQT 136
E D S G EDED SE+ ED S+ D + DS L +QT
Sbjct: 3648 EDEDEPSEKGL-EDEDEPSENSLEDEDEPSENDDYQNRLDSYQDRTGLLDAVQT 3806
Score = 38.5 bits (88), Expect = 0.006
Identities = 32/131 (24%), Positives = 58/131 (43%), Gaps = 13/131 (9%)
Frame = +3
Query: 51 SDSDLSSDGDD---QLADDFLQ----------GSDDEDNDNDNDDEGSDLESGSGSDEDE 97
SD + S DD Q A D L+ SD++ +DND DD+ L+ G+ + E E
Sbjct: 2709 SDKEKDSGNDDDKLQKASDILEPVNYSAENYSDSDNDSDDNDPDDDYDTLQKGTDTLEPE 2888
Query: 98 DSDSESGEEDIARKSKLIDRKRERDSQAAADELQTNIQTNIQDESDEFTLPTKQELEEEA 157
+ + + I R +++ + + D++ DE DE D+ K + +
Sbjct: 2889 NDNDTDDYDKIQRAHYILEPENDSDTE-PDDEPDYEPDDEPDDEPDD---DDKLQKASDI 3056
Query: 158 LRPPDLSNLQR 168
L P + S+ ++
Sbjct: 3057 LEPENPSDSEK 3089
>TC77681 similar to GP|22597156|gb|AAN03465.1 nucleolar histone deacetylase
HD2-P39 {Glycine max}, partial (47%)
Length = 1233
Score = 44.7 bits (104), Expect = 9e-05
Identities = 30/110 (27%), Positives = 59/110 (53%), Gaps = 7/110 (6%)
Frame = +2
Query: 25 VPIKDDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEG 84
+P+ + E +A L+ E +D+ ++ + ++ D +D D+D+DDE
Sbjct: 419 IPLITTEQNGKPEPKAEELKVSEPKKADAKNAAPAKNAATAKHVKVVDPKDEDSDDDDE- 595
Query: 85 SDLESGSGSDEDEDSDSESGEEDIARKS-------KLIDRKRERDSQAAA 127
SD E GS DE E++DS+S +ED + + K +D+ ++R +++A+
Sbjct: 596 SDDEIGSSDDEMENADSDSEDEDDSDEDDEEETPVKKVDQGKKRPNESAS 745
Score = 36.6 bits (83), Expect = 0.024
Identities = 23/101 (22%), Positives = 46/101 (44%)
Frame = +2
Query: 3 PATKKKPSKPVKPQKAPKKVQIVPIKDDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQ 62
P K + K +P+KA K P K+ +T++ + P + + D D DD+
Sbjct: 452 PEPKAEELKVSEPKKADAK-NAAPAKNAATAKHVKVVDPKDEDSDDDDESDDEIGSSDDE 628
Query: 63 LADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSES 103
+ + D++D+D D+++E + G +S S++
Sbjct: 629 MENADSDSEDEDDSDEDDEEETPVKKVDQGKKRPNESASKT 751
>TC76523 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (69%)
Length = 1615
Score = 43.9 bits (102), Expect = 1e-04
Identities = 29/100 (29%), Positives = 46/100 (46%)
Frame = +3
Query: 7 KKPSKPVKPQKAPKKVQIVPIKDDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLADD 66
K SKP K KK +D S S+E++ P E+ S+SD S D D D
Sbjct: 213 KPVSKPAAVAKKSKKDSSDSDDEDDDSSSDEDKKPVAAKKEVSESESDSSDDEDKMNVDK 392
Query: 67 FLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEE 106
GSD ++++ +++DE S D + +SG++
Sbjct: 393 --DGSDSDESEEESEDEPSKTPQKKIKDVEMVDAGKSGKK 506
Score = 39.3 bits (90), Expect = 0.004
Identities = 30/119 (25%), Positives = 55/119 (46%), Gaps = 13/119 (10%)
Frame = +3
Query: 11 KPVKPQKAPKKVQIVPIKDDSTSESEEEE-APPLQH------------LELDGSDSDLSS 57
KP+K P++ ++S ES+E+E A P+ + D D D SS
Sbjct: 120 KPLKM*VHPQRKPASSSDEESDEESDEDEDAKPVSKPAAVAKKSKKDSSDSDDEDDDSSS 299
Query: 58 DGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEEDIARKSKLID 116
D D + + S+ E + +D++D+ + + GS SDE E+ + + +K K ++
Sbjct: 300 DEDKKPVAAKKEVSESESDSSDDEDKMNVDKDGSDSDESEEESEDEPSKTPQKKIKDVE 476
Score = 33.5 bits (75), Expect = 0.20
Identities = 38/161 (23%), Positives = 75/161 (45%)
Frame = +3
Query: 16 QKAPKKVQIVPIKDDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDED 75
Q+ P K+ + P + ++S EE + + SD D + + A + S +
Sbjct: 114 QQKPLKM*VHPQRKPASSSDEESD---------EESDEDEDAKPVSKPAA-VAKKSKKDS 263
Query: 76 NDNDNDDEGSDLESGSGSDEDEDSDSESGEEDIARKSKLIDRKRERDSQAAADELQTNIQ 135
+D+D+ ED+DS S+ ++ +A K ++ E +S ++ DE + N+
Sbjct: 264 SDSDD--------------EDDDSSSDEDKKPVAAKKEV----SESESDSSDDEDKMNVD 389
Query: 136 TNIQDESDEFTLPTKQELEEEALRPPDLSNLQRRIKEIVRV 176
+ D SDE +++E E+E + P Q++IK++ V
Sbjct: 390 KDGSD-SDE----SEEESEDEPSKTP-----QKKIKDVEMV 482
>TC91581 weakly similar to GP|7269000|emb|CAB80733.1 contains EST
gb:AI998867.1 {Arabidopsis thaliana}, partial (5%)
Length = 666
Score = 43.5 bits (101), Expect = 2e-04
Identities = 34/118 (28%), Positives = 55/118 (45%), Gaps = 6/118 (5%)
Frame = +2
Query: 6 KKKPSKPVKPQKAPKKVQI-----VPIKDDSTSESEEEEAPPLQHLELDGSDSDLSSDGD 60
++K + P P K++Q V + D + E EE P + + +S + D
Sbjct: 149 QRKKTGPRLPSSLKKEIQHLNPTPVDVDDIDSDVYEYEEEQPEEESRKNKRYDPVSVNDD 328
Query: 61 DQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDE-DSDSESGEEDIARKSKLIDR 117
+ L+ DF +DE+ +D+DD G D G E DSD + GEED R +++ R
Sbjct: 329 NDLSSDF----EDENVQSDDDDGGYDF---IGKKRKEIDSDDDYGEEDDERHERMLQR 481
>TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {Arabidopsis
thaliana}, partial (39%)
Length = 883
Score = 43.5 bits (101), Expect = 2e-04
Identities = 23/86 (26%), Positives = 44/86 (50%), Gaps = 7/86 (8%)
Frame = +3
Query: 28 KDDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLAD-------DFLQGSDDEDNDNDN 80
K D+ + ++++ +Q + DG + D S D ++ D + GSDD + D+
Sbjct: 312 KSDTEDDEDDDDDDDVQDEDDDGEEEDYSGDEGEEEGDPEDDPEANGAGGSDDGE---DD 482
Query: 81 DDEGSDLESGSGSDEDEDSDSESGEE 106
DD+G + + G DE+++ D E +E
Sbjct: 483 DDDGDEEDEEDGEDEEDEEDEEEDDE 560
Score = 39.3 bits (90), Expect = 0.004
Identities = 26/86 (30%), Positives = 44/86 (50%)
Frame = +3
Query: 28 KDDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDL 87
++D + + EEE P E +G+ S DG+D DD+D D +++++G D
Sbjct: 384 EEDYSGDEGEEEGDPEDDPEANGAGG--SDDGED----------DDDDGDEEDEEDGEDE 527
Query: 88 ESGSGSDEDEDSDSESGEEDIARKSK 113
E +EDE+ D E+ + A+K K
Sbjct: 528 E----DEEDEEEDDETPQPP-AKKRK 590
Score = 31.6 bits (70), Expect = 0.76
Identities = 26/92 (28%), Positives = 45/92 (48%), Gaps = 2/92 (2%)
Frame = +3
Query: 72 DDEDNDN--DNDDEGSDLESGSGSDEDEDSDSESGEEDIARKSKLIDRKRERDSQAAADE 129
DD+D+D+ D DD+G + E SG + +E+ D E E A + D + D ++
Sbjct: 336 DDDDDDDVQDEDDDGEE-EDYSGDEGEEEGDPEDDPE--ANGAGGSDDGEDDDDDGDEED 506
Query: 130 LQTNIQTNIQDESDEFTLPTKQELEEEALRPP 161
+ + +DE DE +E ++E +PP
Sbjct: 507 -----EEDGEDEEDE----EDEEEDDETPQPP 575
>TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~unknown
protein {Arabidopsis thaliana}, partial (46%)
Length = 2077
Score = 43.5 bits (101), Expect = 2e-04
Identities = 27/108 (25%), Positives = 50/108 (46%)
Frame = +2
Query: 29 DDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLE 88
D+ T E+EE + E D +D D D + + A +F +D+D+D++ D+E D+
Sbjct: 593 DELTEYLEKEEDNYEKGEERDEADEDSEEDDELEKAGEFEMDDEDDDDDDEEDEEAEDM- 769
Query: 89 SGSGSDEDEDSDSESGEEDIARKSKLIDRKRERDSQAAADELQTNIQT 136
G+ ED E+ RK +L++ + D + + + T
Sbjct: 770 ---GNVRYEDFFGGKNEKGSKRKDQLLEVSGDEDDMESTKQKKRTAST 904
Score = 43.5 bits (101), Expect = 2e-04
Identities = 51/206 (24%), Positives = 86/206 (40%), Gaps = 22/206 (10%)
Frame = +2
Query: 17 KAPKKVQIVPIKDDSTSESEEEEAPPLQHLELDGSDSDLSSDGDD-------------QL 63
K P+++ + D + E+++ L+ E D D +L D DD +
Sbjct: 293 KNPEEIAQFKVPLDVGKKLEKKKRVELEEEESDDFDEELDDDDDDFEGVEKKKAKGGSEG 472
Query: 64 ADDFLQGSDDED-----NDNDNDDEGSDLESGSGSDE----DEDSDSESGEEDIARKSKL 114
DDF + DDED +++D +DE ++ G D+ DE ++ EED K +
Sbjct: 473 EDDF-EEEDDEDEEGSEDEDDEEDEKEKVKGGGIEDKFLKIDELTEYLEKEEDNYEKGE- 646
Query: 115 IDRKRERDSQAAADELQTNIQTNIQDESDEFTLPTKQELEEEALRPPDLSNLQRRIKEIV 174
+R + DEL+ + + DE D+ E +EEA D+ N V
Sbjct: 647 -ERDEADEDSEEDDELEKAGEFEMDDEDDD----DDDEEDEEA---EDMGN--------V 778
Query: 175 RVLSNFKALRQDGATRKDYVDQLKTD 200
R F + G+ RKD + ++ D
Sbjct: 779 RYEDFFGGKNEKGSKRKDQLLEVSGD 856
>TC90331 weakly similar to GP|21751020|dbj|BAC03887. unnamed protein product
{Homo sapiens}, partial (25%)
Length = 557
Score = 43.5 bits (101), Expect = 2e-04
Identities = 37/113 (32%), Positives = 53/113 (46%), Gaps = 10/113 (8%)
Frame = -1
Query: 49 DGSDSDLSSDGDDQLAD---DFLQ---GSDDEDNDNDND-DEGSDLESGSGSDE---DED 98
DG D D + DD+ D +L G DD ND D D+G + E G +E DED
Sbjct: 533 DGDDDDGEFEDDDEEEDFGAGYLVQPVGQDDPLNDGAADIDDGEENEDGEEEEEEVDDED 354
Query: 99 SDSESGEEDIARKSKLIDRKRERDSQAAADELQTNIQTNIQDESDEFTLPTKQ 151
D + +E + S RKR+ D +A D+ +DE+ FT P+K+
Sbjct: 353 VDDDDAQEVLPPASSHPKRKRDNDDEADEDD---------EDEA-AFTKPSKK 225
Score = 28.9 bits (63), Expect = 5.0
Identities = 12/33 (36%), Positives = 21/33 (63%)
Frame = -1
Query: 72 DDEDNDNDNDDEGSDLESGSGSDEDEDSDSESG 104
++ED+D+D DD+ + E D+DE+ D +G
Sbjct: 554 EEEDDDDDGDDDDGEFE-----DDDEEEDFGAG 471
>TC93070 weakly similar to GP|17065230|gb|AAL32769.1 Unknown protein
{Arabidopsis thaliana}, partial (19%)
Length = 679
Score = 43.1 bits (100), Expect = 3e-04
Identities = 22/67 (32%), Positives = 32/67 (46%)
Frame = +1
Query: 58 DGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEEDIARKSKLIDR 117
D DD++ + + +DED D D DD+ D DED+D D + D+ K K +
Sbjct: 220 DSDDEIEEMDFEDDEDEDEDEDEDDDDDD----DDDDEDDDDDGFAEPTDLNAKDKRLKS 387
Query: 118 KRERDSQ 124
K D Q
Sbjct: 388 KTVPDRQ 408
Score = 36.2 bits (82), Expect = 0.031
Identities = 35/123 (28%), Positives = 51/123 (41%), Gaps = 11/123 (8%)
Frame = +1
Query: 47 ELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEG-----------SDLESGSGSDE 95
E+D D + + +D+ DD DD+D+D D+DD+G L+S + D
Sbjct: 241 EMDFEDDEDEDEDEDEDDDD-----DDDDDDEDDDDDGFAEPTDLNAKDKRLKSKTVPDR 405
Query: 96 DEDSDSESGEEDIARKSKLIDRKRERDSQAAADELQTNIQTNIQDESDEFTLPTKQELEE 155
++ + E G + KR SQ A LQ N + S E P EL E
Sbjct: 406 QQEKEKEKGVRSLNNGQS----KRLPKSQRIA-SLQENNSAKFRRNSMEKKYP---ELSE 561
Query: 156 EAL 158
E L
Sbjct: 562 EIL 570
Score = 33.1 bits (74), Expect = 0.26
Identities = 23/83 (27%), Positives = 41/83 (48%), Gaps = 1/83 (1%)
Frame = +1
Query: 49 DGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEEDI 108
D D D +D +D + DD+D+D+D+D++ D G E D ++ +D
Sbjct: 220 DSDDEIEEMDFEDDEDEDEDEDEDDDDDDDDDDEDDDD----DGFAEPTDLNA----KDK 375
Query: 109 ARKSKLI-DRKRERDSQAAADEL 130
KSK + DR++E++ + L
Sbjct: 376 RLKSKTVPDRQQEKEKEKGVRSL 444
>TC77438 homologue to PIR|T06807|T06807 nucleosome assembly protein 1 -
garden pea, partial (93%)
Length = 1607
Score = 43.1 bits (100), Expect = 3e-04
Identities = 37/131 (28%), Positives = 57/131 (43%), Gaps = 18/131 (13%)
Frame = +2
Query: 6 KKKPSKPVKPQKAPKKVQIV---------PIKDDSTSESEEEEAPPLQHLELDGSDSDLS 56
KKKP K K K K + P + + +E+ A LQ+ +++ D D+
Sbjct: 779 KKKPKKGAKNAKPITKTETCESFFNFFNPPEVPEDDEDIDEDMAEELQN-QME-QDYDIG 952
Query: 57 SDGDDQL--------ADDFLQGSDDEDNDNDNDDEGSDLESGS-GSDEDEDSDSESGEED 107
S D++ + QG + D D+++DDE D E DEDED D + EE+
Sbjct: 953 STIRDKIIPHAVSWFTGEAAQGEEFGDLDDEDDDEDDDAEDDDEDEDEDEDEDDDEDEEE 1132
Query: 108 IARKSKLIDRK 118
K K +K
Sbjct: 1133TKTKKKSSAKK 1165
Score = 32.0 bits (71), Expect = 0.59
Identities = 16/57 (28%), Positives = 26/57 (45%)
Frame = +2
Query: 50 GSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEE 106
G D D DD DD +DED D+D D+E + + S + + + G++
Sbjct: 1028 GDLDDEDDDEDDDAEDDDEDEDEDEDEDDDEDEEETKTKKKSSAKKSGIAQLGEGQQ 1198
>BF641220 weakly similar to PIR|G86203|G86 probable N-arginine dibasic
convertase [imported] - Arabidopsis thaliana, partial
(5%)
Length = 634
Score = 42.7 bits (99), Expect = 3e-04
Identities = 25/66 (37%), Positives = 35/66 (52%), Gaps = 1/66 (1%)
Frame = +2
Query: 49 DGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDS-DSESGEED 107
+G+ D S D DD+ DD +DE++D ++DDEG D E DEDE G +
Sbjct: 332 EGAPKDGSIDEDDEEEDD-----EDEEDDEEDDDEGEDDEDEEXEDEDEXXVXGREGGKG 496
Query: 108 IARKSK 113
A +SK
Sbjct: 497 AANQSK 514
Score = 32.7 bits (73), Expect = 0.34
Identities = 19/58 (32%), Positives = 30/58 (50%), Gaps = 2/58 (3%)
Frame = +2
Query: 38 EEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEG--SDLESGSGS 93
E AP ++ D + D + DD+ DD +G DDED + +++DE E G G+
Sbjct: 332 EGAPKDGSIDEDDEEEDDEDEEDDEEDDD--EGEDDEDEEXEDEDEXXVXGREGGKGA 499
>TC83157 similar to GP|21595368|gb|AAM66095.1 unknown {Arabidopsis
thaliana}, partial (57%)
Length = 1108
Score = 42.7 bits (99), Expect = 3e-04
Identities = 24/86 (27%), Positives = 44/86 (50%)
Frame = +3
Query: 52 DSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEEDIARK 111
D + G ++ D+ DD D D+D+DD+ D E G DED+D+ + S +RK
Sbjct: 303 DDHIKEGGTHEIEDE----DDDSDGDDDDDDDDYDEEEGDYEDEDDDTVAVS-----SRK 455
Query: 112 SKLIDRKRERDSQAAADELQTNIQTN 137
+ ++E +++A ++ +Q N
Sbjct: 456 KVYTEEEKEAEAEAIGYKVVGPLQKN 533
>BQ140761 weakly similar to GP|18076245|emb phosphophoryn {Rattus
norvegicus}, partial (9%)
Length = 620
Score = 42.4 bits (98), Expect = 4e-04
Identities = 31/131 (23%), Positives = 52/131 (39%), Gaps = 18/131 (13%)
Frame = +1
Query: 31 STSESEEEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLES- 89
+T ++ + +A + D SDSD S D +D D+ +D+ +DD SD E
Sbjct: 196 ATKQAAQSDAMDVDEKSSDSSDSDSSDSSDGSDSDSDASDDSDDSSDSSSDDSSSDEEPV 375
Query: 90 -----------------GSGSDEDEDSDSESGEEDIARKSKLIDRKRERDSQAAADELQT 132
S SD +DS SES E + + DS +A+ + +
Sbjct: 376 PAPKKEKAKKAKKAKSVSSSSDSSDDSSSESESEP---EPXNVPLPESDDSSSASSDSDS 546
Query: 133 NIQTNIQDESD 143
+ ++ SD
Sbjct: 547 SSDSSSDSSSD 579
Score = 42.0 bits (97), Expect = 6e-04
Identities = 35/101 (34%), Positives = 40/101 (38%)
Frame = +1
Query: 3 PATKKKPSKPVKPQKAPKKVQIVPIKDDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQ 62
PA KK+ +K K +KA DDS+SESE E P L S SSD D
Sbjct: 376 PAPKKEKAK--KAKKAKSVSSSSDSSDDSSSESESEPEPXNVPLPESDDSSSASSDSD-- 543
Query: 63 LADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSES 103
+ D SD S S S D DS SES
Sbjct: 544 ----------------SSSDSSSDSSSDSSSXSDXDSSSES 618
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.313 0.133 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,122,043
Number of Sequences: 36976
Number of extensions: 144935
Number of successful extensions: 2125
Number of sequences better than 10.0: 225
Number of HSP's better than 10.0 without gapping: 1109
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1518
length of query: 513
length of database: 9,014,727
effective HSP length: 100
effective length of query: 413
effective length of database: 5,317,127
effective search space: 2195973451
effective search space used: 2195973451
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146751.12


