

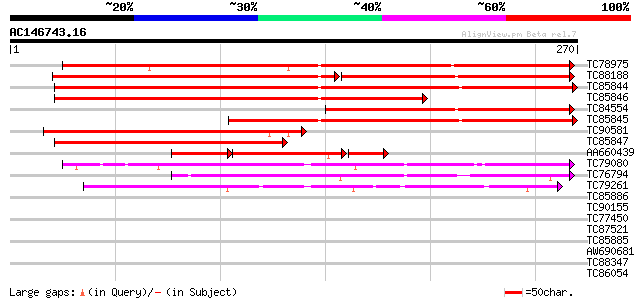
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146743.16 - phase: 2 /pseudo
(270 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC78975 similar to GP|21592656|gb|AAM64605.1 alcohol dehydrogena... 225 1e-59
TC88188 homologue to SP|P80572|ADHX_PEA Alcohol dehydrogenase cl... 142 3e-59
TC85844 homologue to SP|P12886|ADH1_PEA Alcohol dehydrogenase 1 ... 218 1e-57
TC85846 homologue to SP|P12886|ADH1_PEA Alcohol dehydrogenase 1 ... 167 3e-42
TC84554 weakly similar to GP|7705214|gb|AAB33480.2| alcohol dehy... 152 2e-37
TC85845 homologue to SP|P12886|ADH1_PEA Alcohol dehydrogenase 1 ... 142 2e-34
TC90581 similar to PIR|E86357|E86357 alcohol dehydrogenase homol... 134 3e-32
TC85847 homologue to SP|P13603|ADH1_TRIRP Alcohol dehydrogenase ... 116 1e-26
AA660439 similar to PIR|S52036|S520 probable alcohol dehydrogena... 65 1e-24
TC79080 similar to GP|10177043|dbj|BAB10455. alcohol dehydrogena... 99 2e-21
TC76794 similar to GP|7416846|dbj|BAA94084.1 NAD-dependent sorbi... 64 6e-11
TC79261 similar to PIR|T49047|T49047 quinone reductase-like prot... 46 2e-05
TC85886 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydro... 40 0.001
TC90155 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydro... 37 0.008
TC77450 similar to PIR|D86188|D86188 hypothetical protein [impor... 35 0.030
TC87521 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydro... 35 0.040
TC85885 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydro... 31 0.44
AW690681 similar to PIR|D86188|D86 hypothetical protein [importe... 30 0.75
TC88347 homologue to GP|13774427|gb|AAK38870.1 translation initi... 30 1.3
TC86054 beta-amyrin synthase [Medicago truncatula] 29 2.2
>TC78975 similar to GP|21592656|gb|AAM64605.1 alcohol dehydrogenase
putative {Arabidopsis thaliana}, partial (77%)
Length = 1691
Score = 225 bits (574), Expect = 1e-59
Identities = 118/247 (47%), Positives = 169/247 (67%), Gaps = 3/247 (1%)
Frame = +2
Query: 26 NGNTRFTDLNGEIIYHFMFISSFSEYTVVDIANVLKIDPQ--IPPHRACLLGCGVSTGVG 83
+G +RF+ ++G+ I+HF+ S+F+EYTVVD A +K++ + + + LL CGVSTG+G
Sbjct: 485 DGTSRFSSIDGKPIFHFLNTSTFTEYTVVDSACAVKLNTEDNLSLKKLTLLSCGVSTGIG 664
Query: 84 AAWRTAGVEPGSSVAIFGLGSVGLAVAEGARLCGATRIIGVDVNHEKF-EIGKKFGLTDF 142
AAW A V GSSVAIFGLG+VGLAVAEGAR GA++IIGVD+N +KF + G+T+F
Sbjct: 665 AAWNNANVHAGSSVAIFGLGAVGLAVAEGARARGASKIIGVDINPDKFNNTAETMGITEF 844
Query: 143 VHGEECGNKSVSQIIIEMTDGGADYCFECVGMASLVHEAYASCRKGWGKTIVLGVDKPGA 202
++ ++ K V +II EMTDGG DY FEC G +++ +++ S +GWG T++LG+
Sbjct: 845 INPKD-EEKPVYEIIREMTDGGVDYSFECTGNLNVLRDSFLSVHEGWGLTVLLGIHGSPK 1021
Query: 203 RLSLSSSEVLHDGKSLMGSVYGGLKPKSHVPILLKRYMDKELQLDEFVTHELEFKDINKA 262
L + E L DG+ + GSV+GG K KS +P L M ++LD F+THEL F +IN+A
Sbjct: 1022LLPIHPME-LFDGRRIEGSVFGGFKGKSQLPNLATECMKGAIKLDNFITHELPFDEINQA 1198
Query: 263 FDLLSKG 269
F+LL G
Sbjct: 1199FNLLIAG 1219
>TC88188 homologue to SP|P80572|ADHX_PEA Alcohol dehydrogenase class III (EC
1.1.1.1) (Glutathione-dependent formaldehyde
dehydrogenase), complete
Length = 1597
Score = 142 bits (358), Expect(2) = 3e-59
Identities = 71/137 (51%), Positives = 95/137 (68%)
Frame = +1
Query: 21 LGLPRNGNTRFTDLNGEIIYHFMFISSFSEYTVVDIANVLKIDPQIPPHRACLLGCGVST 80
+G+ N + G+ IYHFM S+FS+YTVV +V KI P + CLLGCGV T
Sbjct: 592 VGVMMNDRKPRFSIKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCGVPT 771
Query: 81 GVGAAWRTAGVEPGSSVAIFGLGSVGLAVAEGARLCGATRIIGVDVNHEKFEIGKKFGLT 140
G+GA W TA VEPGS VAIFGLG+VGLAVAEGA+ GA+RIIG+D++ KF+ K FG+T
Sbjct: 772 GLGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNFGVT 951
Query: 141 DFVHGEECGNKSVSQII 157
+F++ ++ K + Q+I
Sbjct: 952 EFINPKD-HEKPIQQVI 999
Score = 103 bits (258), Expect(2) = 3e-59
Identities = 46/111 (41%), Positives = 74/111 (66%)
Frame = +2
Query: 159 EMTDGGADYCFECVGMASLVHEAYASCRKGWGKTIVLGVDKPGARLSLSSSEVLHDGKSL 218
++TDGG DY FEC+G S++ A KGWG ++V+GV G +S +++ G+
Sbjct: 1004 DLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVAASGQEISTRPFQLV-TGRVW 1180
Query: 219 MGSVYGGLKPKSHVPILLKRYMDKELQLDEFVTHELEFKDINKAFDLLSKG 269
G+ +GG K +S VP L+++Y+ KE+++DE+VTH L +IN+AFDL+ +G
Sbjct: 1181 KGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEINEAFDLMHEG 1333
>TC85844 homologue to SP|P12886|ADH1_PEA Alcohol dehydrogenase 1 (EC
1.1.1.1). [Garden pea] {Pisum sativum}, complete
Length = 1502
Score = 218 bits (556), Expect = 1e-57
Identities = 110/249 (44%), Positives = 161/249 (64%)
Frame = +2
Query: 22 GLPRNGNTRFTDLNGEIIYHFMFISSFSEYTVVDIANVLKIDPQIPPHRACLLGCGVSTG 81
G+ N N L G+ I+HF+ S+FSEYTVV V KI+P P + C+L CG+ TG
Sbjct: 458 GVMINDNQSRFSLKGQPIHHFVGTSTFSEYTVVHAGCVAKINPDAPLDKVCILSCGICTG 637
Query: 82 VGAAWRTAGVEPGSSVAIFGLGSVGLAVAEGARLCGATRIIGVDVNHEKFEIGKKFGLTD 141
+GA A +PGSSVAIFGLG+VGLA AEGAR+ GA+RIIGVD+ +FE+ KKFG+ +
Sbjct: 638 LGATINVAKPKPGSSVAIFGLGAVGLAAAEGARISGASRIIGVDLVSSRFELAKKFGVNE 817
Query: 142 FVHGEECGNKSVSQIIIEMTDGGADYCFECVGMASLVHEAYASCRKGWGKTIVLGVDKPG 201
FV+ ++ +K V Q+I EMT+GG D EC G + A+ GWG +++GV
Sbjct: 818 FVNPKD-HDKPVQQVIAEMTNGGVDRAVECTGSIQAMISAFECVHDGWGVAVLVGVPNKD 994
Query: 202 ARLSLSSSEVLHDGKSLMGSVYGGLKPKSHVPILLKRYMDKELQLDEFVTHELEFKDINK 261
+L++ ++L G+ YG KP++ +P ++++YM EL+L++F+TH + F +INK
Sbjct: 995 DAFKTHPMNLLNE-RTLKGTFYGNYKPRTDLPNVVEKYMKGELELEKFITHTIPFSEINK 1171
Query: 262 AFDLLSKGD 270
AFD + KG+
Sbjct: 1172AFDYMLKGE 1198
>TC85846 homologue to SP|P12886|ADH1_PEA Alcohol dehydrogenase 1 (EC
1.1.1.1). [Garden pea] {Pisum sativum}, partial (78%)
Length = 955
Score = 167 bits (424), Expect = 3e-42
Identities = 85/178 (47%), Positives = 116/178 (64%)
Frame = +3
Query: 22 GLPRNGNTRFTDLNGEIIYHFMFISSFSEYTVVDIANVLKIDPQIPPHRACLLGCGVSTG 81
G+ N N + G+ I+HF+ S+FSEYTVV V KI+P P + C+L CG+ TG
Sbjct: 414 GVMINDNQSRFSIKGQPIHHFVGTSTFSEYTVVHAGCVAKINPDAPLDKVCILSCGICTG 593
Query: 82 VGAAWRTAGVEPGSSVAIFGLGSVGLAVAEGARLCGATRIIGVDVNHEKFEIGKKFGLTD 141
+GA A +PGSSVAIFGLG+VGLA AEGAR+ GA+RIIGVD+ +FE+ KKFG+ +
Sbjct: 594 LGATINVAKPKPGSSVAIFGLGAVGLAAAEGARISGASRIIGVDLVSSRFELAKKFGVNE 773
Query: 142 FVHGEECGNKSVSQIIIEMTDGGADYCFECVGMASLVHEAYASCRKGWGKTIVLGVDK 199
FV+ ++ +K V Q+I EMT+GG D EC G + A+ GWG +++GV K
Sbjct: 774 FVNPKD-HDKPVQQVIAEMTNGGVDRAVECTGSIQAMISAFECVHDGWGVAVLVGVPK 944
>TC84554 weakly similar to GP|7705214|gb|AAB33480.2| alcohol dehydrogenase
ADH {Lycopersicon esculentum}, partial (27%)
Length = 550
Score = 152 bits (383), Expect = 2e-37
Identities = 68/119 (57%), Positives = 100/119 (83%)
Frame = +2
Query: 151 KSVSQIIIEMTDGGADYCFECVGMASLVHEAYASCRKGWGKTIVLGVDKPGARLSLSSSE 210
KSVS++I +MT+GGADYCFEC+G+ASL+ EA+ S R+GWGKT+++GV+ G+ L+L+ +
Sbjct: 29 KSVSEVIKDMTNGGADYCFECIGLASLMTEAFNSSREGWGKTVIIGVEMHGSPLTLNPYD 208
Query: 211 VLHDGKSLMGSVYGGLKPKSHVPILLKRYMDKELQLDEFVTHELEFKDINKAFDLLSKG 269
+L GK++ GS++GGLKPKS +P+L ++Y+DKEL LD F++ E++FKDINKAFD L +G
Sbjct: 209 IL-KGKTITGSLFGGLKPKSDLPLLAQKYLDKELNLDGFISQEVDFKDINKAFDYLLQG 382
>TC85845 homologue to SP|P12886|ADH1_PEA Alcohol dehydrogenase 1 (EC
1.1.1.1). [Garden pea] {Pisum sativum}, partial (45%)
Length = 734
Score = 142 bits (357), Expect = 2e-34
Identities = 69/166 (41%), Positives = 108/166 (64%)
Frame = +1
Query: 105 VGLAVAEGARLCGATRIIGVDVNHEKFEIGKKFGLTDFVHGEECGNKSVSQIIIEMTDGG 164
VGLA AEGAR+ GA+RIIGVD+ +FE+ KKFG+ +FV+ ++ +K V Q+I EMT+GG
Sbjct: 1 VGLAAAEGARISGASRIIGVDLVSSRFELAKKFGVNEFVNPKD-HDKPVQQVIAEMTNGG 177
Query: 165 ADYCFECVGMASLVHEAYASCRKGWGKTIVLGVDKPGARLSLSSSEVLHDGKSLMGSVYG 224
D EC G + A+ GWG +++GV +L++ ++L G+ YG
Sbjct: 178 VDRAVECTGSIQAMISAFECVHDGWGVAVLVGVPNKDDAFQTHPMNLLNE-RTLKGTFYG 354
Query: 225 GLKPKSHVPILLKRYMDKELQLDEFVTHELEFKDINKAFDLLSKGD 270
KP++ +P ++++YM EL+L++F+TH + F +INKAFD + KG+
Sbjct: 355 NYKPRTDLPNVVEKYMKGELELEKFITHTIPFSEINKAFDYMLKGE 492
>TC90581 similar to PIR|E86357|E86357 alcohol dehydrogenase homolog
[imported] - Arabidopsis thaliana, partial (52%)
Length = 1289
Score = 134 bits (338), Expect = 3e-32
Identities = 67/127 (52%), Positives = 94/127 (73%), Gaps = 2/127 (1%)
Frame = +1
Query: 17 SKFLLGLPRNGNTRFTDLNGEIIYHFMFISSFSEYTVVDIANVLKIDPQIPPHRACLLGC 76
+K + +PR+G +RF D+ GE+++H + +SSFSEYTVVD+ +V+KI IP +ACLL C
Sbjct: 622 NKPIRDMPRDGTSRFRDMKGEVVHHLLGVSSFSEYTVVDVTHVVKITHDIPLDKACLLSC 801
Query: 77 GVSTGVGAAWRTAGVEPGSSVAIFGLGSVGLAVAEGARLCGATRII-GVDVNHEKF-EIG 134
GVSTG+GAAW+ A VE G++VAIFGLG+VGLAVA + GA++I VD+NH++
Sbjct: 802 GVSTGIGAAWKVADVEKGTTVAIFGLGAVGLAVAVATKQRGASKIYRRVDLNHDEI*NEE 981
Query: 135 KKFGLTD 141
K G+TD
Sbjct: 982 NKCGITD 1002
>TC85847 homologue to SP|P13603|ADH1_TRIRP Alcohol dehydrogenase 1 (EC
1.1.1.1). [Creeping white clover] {Trifolium repens},
partial (60%)
Length = 751
Score = 116 bits (290), Expect = 1e-26
Identities = 59/111 (53%), Positives = 76/111 (68%)
Frame = +1
Query: 22 GLPRNGNTRFTDLNGEIIYHFMFISSFSEYTVVDIANVLKIDPQIPPHRACLLGCGVSTG 81
G+ N N + G+ I+HF+ S+FSEYTVV V KI+P P + C+L CG+ TG
Sbjct: 418 GVMINDNQSRFSIKGQPIHHFVGTSTFSEYTVVHAGCVAKINPDAPLDKVCILSCGICTG 597
Query: 82 VGAAWRTAGVEPGSSVAIFGLGSVGLAVAEGARLCGATRIIGVDVNHEKFE 132
+GA A +PGSSVAIFGLG+VGLA AEGAR+ GA+RIIGVD+ +FE
Sbjct: 598 LGATINVAKPKPGSSVAIFGLGAVGLAAAEGARISGASRIIGVDLVSSRFE 750
>AA660439 similar to PIR|S52036|S520 probable alcohol dehydrogenase (EC
1.1.1.1) ADH3b - tomato, partial (21%)
Length = 403
Score = 65.5 bits (158), Expect(3) = 1e-24
Identities = 33/55 (60%), Positives = 44/55 (80%), Gaps = 1/55 (1%)
Frame = +2
Query: 107 LAVAEGARLCGATRIIGVDVNHEKFEIGKKFGLTDFVHGEECGN-KSVSQIIIEM 160
LAVA A+ GA++IIGVD+NH+KFEIGK+FG+TDFV+ N KSVS++I +M
Sbjct: 89 LAVAVAAKQRGASKIIGVDLNHDKFEIGKQFGITDFVNPSSTSNEKSVSEVIKDM 253
Score = 48.1 bits (113), Expect(3) = 1e-24
Identities = 21/29 (72%), Positives = 26/29 (89%)
Frame = +3
Query: 78 VSTGVGAAWRTAGVEPGSSVAIFGLGSVG 106
VSTG+GAAW+ A VE G++VAIFGLG+VG
Sbjct: 3 VSTGIGAAWKVADVEKGTTVAIFGLGAVG 89
Score = 36.6 bits (83), Expect(3) = 1e-24
Identities = 13/19 (68%), Positives = 18/19 (94%)
Frame = +1
Query: 162 DGGADYCFECVGMASLVHE 180
+GGADYCFEC+G+A+L+ E
Sbjct: 256 NGGADYCFECIGLATLMTE 312
>TC79080 similar to GP|10177043|dbj|BAB10455. alcohol dehydrogenase-like
protein {Arabidopsis thaliana}, partial (96%)
Length = 1524
Score = 98.6 bits (244), Expect = 2e-21
Identities = 79/247 (31%), Positives = 122/247 (48%), Gaps = 3/247 (1%)
Frame = +3
Query: 26 NGNTR-FTDLNGEIIYHFMFISSFSEYTVVDIANVLKIDPQIPPH-RACLLGCGVSTGVG 83
+G TR F +G IY + + +EY VV AN L + P P+ + +LGC V T G
Sbjct: 597 DGETRLFLRGSGNPIYMYS-MGGLAEYCVVP-ANALAVLPNSMPYTESAILGCAVFTAYG 770
Query: 84 AAWRTAGVEPGSSVAIFGLGSVGLAVAEGARLCGATRIIGVDVNHEKFEIGKKFGLTDFV 143
A A V PG +VA+ G G VG + + AR GA+ II VDV EK E K G T
Sbjct: 771 AMAHAAEVRPGDTVAVIGTGGVGSSCLQIARAFGASDIIAVDVQDEKLEKAKTLGAT--- 941
Query: 144 HGEECGNKSVSQIIIEMTDG-GADYCFECVGMASLVHEAYASCRKGWGKTIVLGVDKPGA 202
H + + I+E+T G G D E +G + S + G GK +++G+ + G+
Sbjct: 942 HTINSAKEDPIEKILEITGGKGVDVAVEALGRPQTFAQCTQSVKDG-GKAVMIGLAQAGS 1118
Query: 203 RLSLSSSEVLHDGKSLMGSVYGGLKPKSHVPILLKRYMDKELQLDEFVTHELEFKDINKA 262
+ + ++ ++GS YGG + + +P L++ L VT + F++ KA
Sbjct: 1119LGEVDINRLVRRKIKVIGS-YGG-RARQDLPKLIRLAETGIFDLGHAVTRKYTFEESGKA 1292
Query: 263 FDLLSKG 269
F L++G
Sbjct: 1293FQDLNEG 1313
>TC76794 similar to GP|7416846|dbj|BAA94084.1 NAD-dependent sorbitol
dehydrogenase {Prunus persica}, partial (93%)
Length = 1691
Score = 63.9 bits (154), Expect = 6e-11
Identities = 50/196 (25%), Positives = 86/196 (43%), Gaps = 4/196 (2%)
Frame = +2
Query: 78 VSTGVGAAWRTAGVEPGSSVAIFGLGSVGLAVAEGARLCGATRIIGVDVNHEKFEIGKKF 137
+S GV A R A + P ++V I G G +GL AR GA RI+ VDV+ + + K
Sbjct: 707 LSVGVHAC-RRANIGPETNVLIMGAGPIGLVTMLSARAFGAPRIVVVDVDDHRLSVAKSL 883
Query: 138 GLTDFVHGEECGNKSVSQI--IIEMTDGGADYCFECVGMASLVHEAYASCRKGWGKTIVL 195
G D V ++ I + G D F+C G + A + + G GK ++
Sbjct: 884 GADDIVKVSTNIQDVAEEVKQIHNVLGAGVDVTFDCAGFNKTMTTALTATQPG-GKVCLV 1060
Query: 196 GVDKPGARLSLSSSEVLHDGKSLMGSVYGGLKPKSHVPILLKRYMDKELQLDEFVTHELE 255
G+ + L+ + V G + K+ P+ L+ ++ + +TH
Sbjct: 1061GMGHSEMTVPLTPAAARE------VDVVGIFRYKNTWPLCLEFLRSGKIDVKPLITHRFG 1222
Query: 256 F--KDINKAFDLLSKG 269
F K++ +AF+ ++G
Sbjct: 1223FSQKEVEEAFETSARG 1270
>TC79261 similar to PIR|T49047|T49047 quinone reductase-like protein -
Arabidopsis thaliana, partial (95%)
Length = 1239
Score = 45.8 bits (107), Expect = 2e-05
Identities = 54/239 (22%), Positives = 100/239 (41%), Gaps = 11/239 (4%)
Frame = +3
Query: 36 GEIIYHFMFISSFSEYTVVDIANVLKIDPQIPPHRACLLGCGVSTGVGAAWRTAGVEPGS 95
G+ + F + S+++Y VVD + ++ A L T A ++ G
Sbjct: 300 GDPVCSFAALGSYAQYLVVDQTQLFRVPEGCDLVAAGALAVAFGTSHVGLVHRAQLKSGQ 479
Query: 96 SVAIFGL-GSVGLAVAEGARLCGATRIIGVDVNHEKFEIGKKFGLTDFVHGEECGNKSVS 154
+ + G G VGLA + + CGA +I V EK ++ K G+ H + GN++V+
Sbjct: 480 VLLVLGAAGGVGLAAVQIGKACGAI-VIAVARGAEKVQLLKSMGVD---HVVDLGNENVT 647
Query: 155 QIIIEMTD----GGADYCFECVGMASLVHEAYASCRKGWGKTIVLGVDKPGARLSLSSSE 210
+ + E G D ++ VG L+ E+ + WG I++ G + ++
Sbjct: 648 ESVKEFLKVKRLKGIDVLYDPVG-GKLMKESLRLLK--WGANILIIGFASGEIPVIPANI 818
Query: 211 VLHDGKSLMGSVYGGLKPKSHVPILLKRYMDKELQ------LDEFVTHELEFKDINKAF 263
L ++ G +G K H P +L+ + + L + ++H + N AF
Sbjct: 819 ALVKNWTVHGLYWGSY--KIHRPAVLEDSLKELLSWLAKGLISVHISHSYGLSEANLAF 989
>TC85886 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (94%)
Length = 1395
Score = 39.7 bits (91), Expect = 0.001
Identities = 64/249 (25%), Positives = 102/249 (40%), Gaps = 4/249 (1%)
Frame = +2
Query: 26 NGNTRFTDLNGEIIYHFMFISSFSEYTVVDIANVLKIDPQIPPHRACLLGCGVSTGVGAA 85
NG +R +G I Y FS+ V D V++I +P A L C T V +
Sbjct: 443 NGKSR----DGTITY-----GGFSDSMVADEHFVIRIPDSLPLDGAGPLLCAGVT-VYSP 592
Query: 86 WRTAGVE-PGSSVAIFGLGSVGLAVAEGARLCGA-TRIIGVDVNHEKFEIGKKFGLTDFV 143
R G++ PG ++ + GLG +G + A+ GA +I EK E + G F+
Sbjct: 593 LRHFGLDKPGMNIGVVGLGGLGHMAVKFAKAFGAKVTVISTSPKKEK-EAIEHLGADSFL 769
Query: 144 HGEECGNKSVSQIIIEMTDGGADYCFECVGMASLVHEAYASCRKGWGKTIVLG-VDKPGA 202
+ Q +G D + L+ K GK +++G V KP
Sbjct: 770 VSRD---PEQMQAATSTLNGIIDTVSASHPVVPLI-----GLLKSNGKLVMVGAVAKP-- 919
Query: 203 RLSLSSSEVLHDGKSLMGSVYGGLK-PKSHVPILLKRYMDKELQLDEFVTHELEFKDINK 261
L L +L KS+ GS+ GG+K + + K + E+++ + +N
Sbjct: 920 -LELPIFSLLGGRKSIAGSLIGGIKETQEMIDFAAKHNVTPEIEV-------VPIDYVNT 1075
Query: 262 AFDLLSKGD 270
A + L KGD
Sbjct: 1076AMERLVKGD 1102
>TC90155 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (78%)
Length = 1051
Score = 37.0 bits (84), Expect = 0.008
Identities = 54/184 (29%), Positives = 74/184 (39%), Gaps = 4/184 (2%)
Frame = +2
Query: 48 FSEYTVVDIANVLKIDPQIPPHRACLLGCGVSTGVGAAWRTAGVE-PGSSVAIFGLGSVG 106
+S+ V D V++I IP A L C T V + R G++ PG + + GLG +G
Sbjct: 191 YSDSMVADEHFVIRIPDNIPLEFAGPLLCAGVT-VYSPLRFFGLDKPGLHIGVVGLGGLG 367
Query: 107 LAVAEGARLCGA-TRIIGVDVNHEKFEIGKKFGLTDFVHGEECGNKSVSQIIIEMTDGGA 165
+ A+ GA +I N EK E + G F+ + Q I DG
Sbjct: 368 HMAVKFAKAFGANVTVISTSPNKEK-EAIEHLGADSFLISSD---PKQIQGAIGTLDG-- 529
Query: 166 DYCFECVGMASLVHE--AYASCRKGWGKTIVLGVDKPGARLSLSSSEVLHDGKSLMGSVY 223
+ S VH K GK I+LGV L L ++ K L GS
Sbjct: 530 -----IIDTVSAVHPLLPMIGLLKSHGKLIMLGVIV--QPLQLPEYTLIQGRKILAGSQV 688
Query: 224 GGLK 227
GGLK
Sbjct: 689 GGLK 700
>TC77450 similar to PIR|D86188|D86188 hypothetical protein [imported] -
Arabidopsis thaliana, partial (79%)
Length = 1645
Score = 35.0 bits (79), Expect = 0.030
Identities = 32/127 (25%), Positives = 57/127 (44%), Gaps = 13/127 (10%)
Frame = +3
Query: 96 SVAIFGLGSVGLAVAEGARLCGATRIIGVDVNHEKFEIGKKFGLTDFVHGEECGNKSVSQ 155
S+AI G+G VG AE CG R++ D ++K E+ L F ++ G
Sbjct: 354 SIAIVGIGGVGSVAAEMLTRCGIGRLLLYD--YDKVELANMNRL--FFRPDQAGMTKTDA 521
Query: 156 IIIEMTDGGADYCFEC------------VGMASLVHEAYASCRKGWGKTIVLG-VDKPGA 202
+ ++D D E M+SL ++++ ++G G +VL VD A
Sbjct: 522 ALQTLSDINPDVVLESFTLNITTVDGFETFMSSLKNKSFRPDKQGSGVDLVLSCVDNYEA 701
Query: 203 RLSLSSS 209
R++++ +
Sbjct: 702 RMAVNQA 722
>TC87521 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (98%)
Length = 1433
Score = 34.7 bits (78), Expect = 0.040
Identities = 56/210 (26%), Positives = 88/210 (41%), Gaps = 6/210 (2%)
Frame = +2
Query: 24 PRNGNT---RFTDLNGEIIYHFMFISSFSEYTVVDIANVLKIDPQIPPHRACLLGCGVST 80
P+ NT +++D G I Y +S+ V D ++ I +P A L C T
Sbjct: 419 PKQTNTYSAKYSD--GSITY-----GGYSDSMVADEHFIVHIPDGLPLESAAPLLCAGIT 577
Query: 81 GVGAAWRTAGVE-PGSSVAIFGLGSVGLAVAEGARLCGA-TRIIGVDVNHEKFEIGKKFG 138
V + R G++ PG ++ I GLG +G + A+ GA +I N EK E + G
Sbjct: 578 -VYSPLRYFGLDKPGMNIGIVGLGGLGHLGVKFAKAFGANVTVISTSPNKEK-EAIENLG 751
Query: 139 LTDFVHGEECGNKSVSQIIIEMTDGGADYCFECVGMASLVHEAYASCRKGWGKTIVLGV- 197
F+ + + Q + DG D + LV K GK +++G
Sbjct: 752 ADSFLISHD---QDKMQAAMGTLDGIIDTVSADHPLLPLV-----GLLKYHGKLVMVGAP 907
Query: 198 DKPGARLSLSSSEVLHDGKSLMGSVYGGLK 227
DKP L ++ K++ GS GG+K
Sbjct: 908 DKPP---ELPHIPLIMGRKTISGSGIGGMK 988
>TC85885 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (97%)
Length = 1330
Score = 31.2 bits (69), Expect = 0.44
Identities = 50/227 (22%), Positives = 84/227 (36%), Gaps = 4/227 (1%)
Frame = +3
Query: 48 FSEYTVVDIANVLKIDPQIPPHRACLLGCGVSTGVGAAWRTAGVEPGSSVAIFGLGSVGL 107
+S+ V D V++I +P A L C T +PG ++ + GLG +G
Sbjct: 480 YSDSMVADEHFVIRIPDSLPLDVAGPLLCAGVTVYSPLRHFQLDKPGMNIGVVGLGGLGH 659
Query: 108 AVAEGARLCGA-TRIIGVDVNHEKFEIGKKFGLTDFVHGEECGNKSVSQIIIEMTDGGAD 166
+ A+ GA +I + EK E + G F+ + + G +
Sbjct: 660 MAVKFAKAFGANVTVISTSPSKEK-EAIEHLGADSFLVSRDPDQMQAAM-------GTLN 815
Query: 167 YCFECVGMASLVHEAYASCRKGWGKTIVLGVDKPGARLSLSSSEVLHDGKSLMGSVYGGL 226
+ V + + + +V GV KP L L +L K + GS+ GG+
Sbjct: 816 GIIDTVSASHPILPLIGLLKSNGKLVMVGGVAKP---LELPVFSLLGGRKLVAGSLIGGI 986
Query: 227 KPKSHVPILLKRYMDKELQLDEFVTHELEFKDI---NKAFDLLSKGD 270
K + + + VT ++E I N A + L K D
Sbjct: 987 KETQEM---------IDFAAEHNVTPDIEVVPIDYVNTAMERLEKAD 1100
>AW690681 similar to PIR|D86188|D86 hypothetical protein [imported] -
Arabidopsis thaliana, partial (35%)
Length = 591
Score = 30.4 bits (67), Expect = 0.75
Identities = 25/94 (26%), Positives = 41/94 (43%)
Frame = +2
Query: 96 SVAIFGLGSVGLAVAEGARLCGATRIIGVDVNHEKFEIGKKFGLTDFVHGEECGNKSVSQ 155
S+AI G+G VG AE CG R++ D ++K E+ L F ++ G
Sbjct: 281 SIAIVGIGGVGSVAAEMLTRCGIGRLLLYD--YDKVELANMNRL--FFRPDQAGMTKTDA 448
Query: 156 IIIEMTDGGADYCFECVGMASLVHEAYASCRKGW 189
+ ++D D E V + +L Y ++ W
Sbjct: 449 ALQTLSDINPDVVLE-VTVWTLS*AVYIIMKQEW 547
>TC88347 homologue to GP|13774427|gb|AAK38870.1 translation initiation
factor IF1 {Glycine max}, partial (42%)
Length = 765
Score = 29.6 bits (65), Expect = 1.3
Identities = 15/51 (29%), Positives = 26/51 (50%)
Frame = +3
Query: 25 RNGNTRFTDLNGEIIYHFMFISSFSEYTVVDIANVLKIDPQIPPHRACLLG 75
RNG+T+ + N + +F+S + ++I LK+ IPP A +G
Sbjct: 354 RNGSTKVSSQNPSLTACSVFVSIMKTLSFLEIELELKLVDMIPPKDASFIG 506
>TC86054 beta-amyrin synthase [Medicago truncatula]
Length = 2784
Score = 28.9 bits (63), Expect = 2.2
Identities = 14/47 (29%), Positives = 23/47 (48%)
Frame = +1
Query: 188 GWGKTIVLGVDKPGARLSLSSSEVLHDGKSLMGSVYGGLKPKSHVPI 234
GWG++ + K L S S V+H +LMG ++ G + P+
Sbjct: 2116 GWGESYLSSPKKIYVPLEGSRSNVVHTAWALMGLIHAGQAERDPTPL 2256
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.141 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,430,006
Number of Sequences: 36976
Number of extensions: 119609
Number of successful extensions: 511
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 499
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 500
length of query: 270
length of database: 9,014,727
effective HSP length: 95
effective length of query: 175
effective length of database: 5,502,007
effective search space: 962851225
effective search space used: 962851225
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146743.16


