

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146743.14 + phase: 0
(720 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
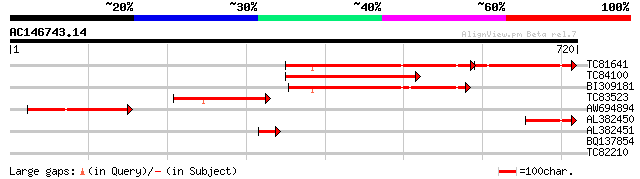
Score E
Sequences producing significant alignments: (bits) Value
TC81641 similar to GP|12313690|dbj|BAB21095. hypothetical protei... 298 e-111
TC84100 similar to PIR|G86208|G86208 protein F22G5.28 [imported]... 351 5e-97
BI309181 similar to PIR|G86208|G86 protein F22G5.28 [imported] -... 278 7e-75
TC83523 similar to PIR|G86208|G86208 protein F22G5.28 [imported]... 244 6e-65
AW694894 similar to GP|12313690|db hypothetical protein {Oryza s... 199 2e-51
AL382450 similar to GP|15982799|gb| AT5g58980/k19m22_180 {Arabid... 59 7e-09
AL382451 similar to PIR|G86208|G862 protein F22G5.28 [imported] ... 45 1e-04
BQ137854 30 4.2
TC82210 weakly similar to GP|15450527|gb|AAK96556.1 AT5g21160/T1... 28 9.5
>TC81641 similar to GP|12313690|dbj|BAB21095. hypothetical protein {Oryza
sativa (japonica cultivar-group)}, partial (41%)
Length = 1249
Score = 298 bits (762), Expect(2) = e-111
Identities = 151/244 (61%), Positives = 180/244 (72%), Gaps = 3/244 (1%)
Frame = +2
Query: 351 ELFGSASEELTGKIDYRHVYLNFTNIEVELDNK---KVVKTCPAALGPGFAAGTTDGPGV 407
ELF ASE++ GK+D+RH YL+F+ +EV + + KVVKTCPAA+G GFAAGTTDGPG
Sbjct: 2 ELFNGASEQIKGKVDFRHAYLDFSKLEVNVSSNGASKVVKTCPAAMGFGFAAGTTDGPGA 181
Query: 408 FGFQQGDPEISPFWKNVRDFLKEPSQYQVDCQNPKPVLLSSGEMFDPYPWAPAILPIQIL 467
F F+QGD + +PFWK VR+ LK P + Q+ CQ PKP+LL +GEM PY WAP ILPIQIL
Sbjct: 182 FDFKQGDDQGNPFWKLVRNLLKTPDKEQIACQQPKPILLDTGEMKLPYDWAPTILPIQIL 361
Query: 468 RLGKLIILSVPGEFTTMAGRRLREAVKETLISNSNGEFNNETHVVIAGLTNTYSQYIATF 527
R+G+ ILSVPGEFTTMAGRRLR+AVK L + + + HVVIAGLTNTYSQY+ T+
Sbjct: 362 RIGQFFILSVPGEFTTMAGRRLRDAVKTVL--SGDKSLGSNIHVVIAGLTNTYSQYVTTY 535
Query: 528 EEYHQQRYEAASTLYGPHTLSAYIQEFNKLAQAMAKGDKIYGNGTSPPDLLSVQKSFLLD 587
EEY QRYE ASTLYGPHTL AYIQEF KLA A+ G + +G PPDLL Q LL
Sbjct: 536 EEYEVQRYEGASTLYGPHTLDAYIQEFKKLAHALINGQPV-ESGPQPPDLLDKQIG-LLT 709
Query: 588 PFGD 591
P D
Sbjct: 710 PSSD 721
Score = 124 bits (312), Expect(2) = e-111
Identities = 66/131 (50%), Positives = 84/131 (63%), Gaps = 2/131 (1%)
Frame = +3
Query: 591 DTTPDGIKLGDIKEDIAFPGSGYFTKGDKPSATFWSANPRYDLLTEGTYAVVERLQG-ER 649
D TP G GD D+ P + F +GD S TFWSA PR DL+TEGT+++VE LQG +
Sbjct: 723 DGTPLGTSFGDCSSDV--PKNSTFKRGDTVSVTFWSACPRNDLMTEGTFSLVEYLQGKDT 896
Query: 650 WISVQDDDDLSLFFRW-KVDNTSFHGFAAIEWEIPTDAISGVYRLKHFGASKKTIVSPIN 708
W+ DDDD + F+W + S H AAIEW IP D GVYR+KHFGA+ K ++ I
Sbjct: 897 WVPAYDDDDFCVRFKWSRPFKLSTHSKAAIEWRIPQDVAPGVYRIKHFGAA-KGLLGSIR 1073
Query: 709 YFTGASSAFAV 719
+FTG+SSAF V
Sbjct: 1074HFTGSSSAFVV 1106
>TC84100 similar to PIR|G86208|G86208 protein F22G5.28 [imported] -
Arabidopsis thaliana, partial (17%)
Length = 516
Score = 351 bits (901), Expect = 5e-97
Identities = 171/171 (100%), Positives = 171/171 (100%)
Frame = +2
Query: 351 ELFGSASEELTGKIDYRHVYLNFTNIEVELDNKKVVKTCPAALGPGFAAGTTDGPGVFGF 410
ELFGSASEELTGKIDYRHVYLNFTNIEVELDNKKVVKTCPAALGPGFAAGTTDGPGVFGF
Sbjct: 2 ELFGSASEELTGKIDYRHVYLNFTNIEVELDNKKVVKTCPAALGPGFAAGTTDGPGVFGF 181
Query: 411 QQGDPEISPFWKNVRDFLKEPSQYQVDCQNPKPVLLSSGEMFDPYPWAPAILPIQILRLG 470
QQGDPEISPFWKNVRDFLKEPSQYQVDCQNPKPVLLSSGEMFDPYPWAPAILPIQILRLG
Sbjct: 182 QQGDPEISPFWKNVRDFLKEPSQYQVDCQNPKPVLLSSGEMFDPYPWAPAILPIQILRLG 361
Query: 471 KLIILSVPGEFTTMAGRRLREAVKETLISNSNGEFNNETHVVIAGLTNTYS 521
KLIILSVPGEFTTMAGRRLREAVKETLISNSNGEFNNETHVVIAGLTNTYS
Sbjct: 362 KLIILSVPGEFTTMAGRRLREAVKETLISNSNGEFNNETHVVIAGLTNTYS 514
>BI309181 similar to PIR|G86208|G86 protein F22G5.28 [imported] - Arabidopsis
thaliana, partial (23%)
Length = 716
Score = 278 bits (710), Expect = 7e-75
Identities = 138/234 (58%), Positives = 179/234 (75%), Gaps = 3/234 (1%)
Frame = +1
Query: 355 SASEELTGKIDYRHVYLNFTNIEVELDNK---KVVKTCPAALGPGFAAGTTDGPGVFGFQ 411
+A EE+ G++D+RH Y++F+ +EV + ++ KVVKTCPAA+G FAAGTTDGPG F F+
Sbjct: 4 AADEEIKGEVDFRHAYIDFSKLEVTISDQGADKVVKTCPAAMGFAFAAGTTDGPGSFDFK 183
Query: 412 QGDPEISPFWKNVRDFLKEPSQYQVDCQNPKPVLLSSGEMFDPYPWAPAILPIQILRLGK 471
QGD + SPFWK VR+ + S+ Q+DCQ+PKP+LL +GEM PY WAP+ILPIQILR+G+
Sbjct: 184 QGDDKGSPFWKLVRNLVLTLSRKQIDCQHPKPILLDTGEMNIPYDWAPSILPIQILRVGQ 363
Query: 472 LIILSVPGEFTTMAGRRLREAVKETLISNSNGEFNNETHVVIAGLTNTYSQYIATFEEYH 531
IILS+PGE +TMAGRRLR+AVK L +S+ +F N H+VI+ L+N YSQY T+EEYH
Sbjct: 364 FIILSIPGEISTMAGRRLRDAVKTVL--SSHKDFEN-VHIVISALSNAYSQYATTYEEYH 534
Query: 532 QQRYEAASTLYGPHTLSAYIQEFNKLAQAMAKGDKIYGNGTSPPDLLSVQKSFL 585
QRYE ASTLYGPHTL+AYIQEF KLA+A+ G + +G PP+LL Q L
Sbjct: 535 VQRYEGASTLYGPHTLNAYIQEFKKLAKALVSGQPV-ESGPQPPNLLDKQVCLL 693
>TC83523 similar to PIR|G86208|G86208 protein F22G5.28 [imported] -
Arabidopsis thaliana, partial (12%)
Length = 455
Score = 244 bits (624), Expect = 6e-65
Identities = 124/151 (82%), Positives = 124/151 (82%), Gaps = 27/151 (17%)
Frame = +2
Query: 208 LKFVDSASGKSKGSFSWFATHGTSMSNNNKLISGDNK----------------------- 244
LKFVDSASGKSKGSFSWFATHGTSMSNNNKLISGDNK
Sbjct: 2 LKFVDSASGKSKGSFSWFATHGTSMSNNNKLISGDNKGVAARLFEDWFTSQNKSSSPNSN 181
Query: 245 ----DIGELVQIAQLIKATGGKDCNEKSSQASKVRKNDGSLFVGAFCQSNVGDVSPNVLG 300
DIGELVQIAQLIKATGGKDCNEKSSQASKVRKNDGSLFVGAFCQSNVGDVSPNVLG
Sbjct: 182 STELDIGELVQIAQLIKATGGKDCNEKSSQASKVRKNDGSLFVGAFCQSNVGDVSPNVLG 361
Query: 301 AFCIDSGKPCDFNHSSCNGNDLLCVGRGPGY 331
AFCIDSGKPCDFNHSSCNGNDLLCVGRGPGY
Sbjct: 362 AFCIDSGKPCDFNHSSCNGNDLLCVGRGPGY 454
>AW694894 similar to GP|12313690|db hypothetical protein {Oryza sativa
(japonica cultivar-group)}, partial (15%)
Length = 665
Score = 199 bits (507), Expect = 2e-51
Identities = 98/133 (73%), Positives = 116/133 (86%)
Frame = +2
Query: 23 YGEYLIGVGSYDMTGPAADVNMMGYANIEQNTAGIHFRLRARTFIVAENLQGPRFVFVNL 82
Y YL+G+GSYD+TGPAADVNMMGYAN EQ +G+HFRLR+R FIVAE +G R VFVNL
Sbjct: 269 YCNYLVGLGSYDITGPAADVNMMGYANTEQIASGVHFRLRSRAFIVAEP-KGNRLVFVNL 445
Query: 83 DAGMASQLLTIKLLERLKSRFGNLYTEENVAISGIHTHAGPGGYLQYVVYSVTSLGFVTQ 142
DA M +QL+TIK+LERLK+R+G++YTE NVAISGIHTHAGPGGYLQYVVY VTSLGFV Q
Sbjct: 446 DACMGAQLVTIKVLERLKARYGDVYTENNVAISGIHTHAGPGGYLQYVVYIVTSLGFVRQ 625
Query: 143 SFDAIANAVEQSI 155
SFDA+ + +E+SI
Sbjct: 626 SFDALVDGIEKSI 664
>AL382450 similar to GP|15982799|gb| AT5g58980/k19m22_180 {Arabidopsis
thaliana}, partial (20%)
Length = 368
Score = 58.9 bits (141), Expect = 7e-09
Identities = 32/66 (48%), Positives = 40/66 (60%), Gaps = 1/66 (1%)
Frame = -3
Query: 655 DDDDLSLFFRW-KVDNTSFHGFAAIEWEIPTDAISGVYRLKHFGASKKTIVSPINYFTGA 713
DDDD L F+W + S A +EW IP GVYR+ HFGA+ K + IN+FTG+
Sbjct: 366 DDDDFCLRFKWSRRFQLSPMSKATMEWRIPQGVTPGVYRISHFGAA-KGLFGSINHFTGS 190
Query: 714 SSAFAV 719
SSAF V
Sbjct: 189 SSAFVV 172
>AL382451 similar to PIR|G86208|G862 protein F22G5.28 [imported] -
Arabidopsis thaliana, partial (3%)
Length = 509
Score = 44.7 bits (104), Expect = 1e-04
Identities = 18/29 (62%), Positives = 23/29 (79%)
Frame = -3
Query: 316 SCNGNDLLCVGRGPGYPNEILSTKIIGER 344
+C G + LC G+GPGYP+E ST+IIGER
Sbjct: 87 TCGGRNELCYGQGPGYPDEFESTRIIGER 1
>BQ137854
Length = 960
Score = 29.6 bits (65), Expect = 4.2
Identities = 20/54 (37%), Positives = 26/54 (48%), Gaps = 4/54 (7%)
Frame = +1
Query: 498 ISNSNGEFNNETHVVIAGLTNT----YSQYIATFEEYHQQRYEAASTLYGPHTL 547
IS S N+E VV + NT S+ AT YH+ ++YGPHTL
Sbjct: 352 ISMSQNSVNHEQKVVQSSAANTRDNTLSRTRATKCIYHRSLERMT*SMYGPHTL 513
>TC82210 weakly similar to GP|15450527|gb|AAK96556.1 AT5g21160/T10F18_190
{Arabidopsis thaliana}, partial (24%)
Length = 1436
Score = 28.5 bits (62), Expect = 9.5
Identities = 13/32 (40%), Positives = 19/32 (58%)
Frame = -3
Query: 268 SSQASKVRKNDGSLFVGAFCQSNVGDVSPNVL 299
SS +S + + S+F G FC S++ S NVL
Sbjct: 819 SSSSSILPVEESSVFFGIFCSSSISSSSKNVL 724
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,801,858
Number of Sequences: 36976
Number of extensions: 276759
Number of successful extensions: 1300
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 1269
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1289
length of query: 720
length of database: 9,014,727
effective HSP length: 103
effective length of query: 617
effective length of database: 5,206,199
effective search space: 3212224783
effective search space used: 3212224783
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146743.14


