

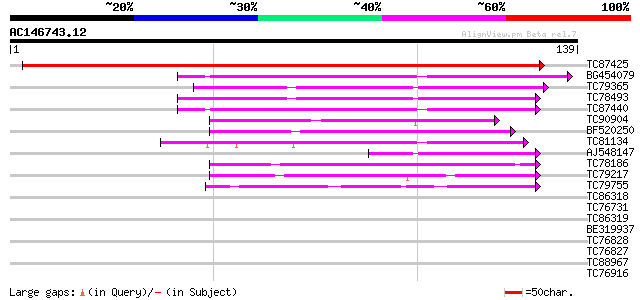
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146743.12 + phase: 0
(139 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC87425 weakly similar to GP|20161478|dbj|BAB90402. contains EST... 259 2e-70
BG454079 similar to GP|10176956|dbj contains similarity to nonsp... 81 1e-16
TC79365 weakly similar to GP|15010698|gb|AAK74008.1 AT3g22600/F1... 78 1e-15
TC78493 weakly similar to GP|15010698|gb|AAK74008.1 AT3g22600/F1... 76 4e-15
TC87440 similar to GP|17979494|gb|AAL50083.1 AT5g64080/MHJ24_6 {... 73 3e-14
TC90904 similar to GP|3128176|gb|AAC16080.1| unknown protein {Ar... 56 5e-09
BF520250 weakly similar to GP|3128176|gb| unknown protein {Arabi... 47 2e-06
TC81134 weakly similar to GP|14334664|gb|AAK59510.1 unknown prot... 43 5e-05
AJ548147 similar to GP|15010698|gb| AT3g22600/F16J14_17 {Arabido... 43 5e-05
TC78186 weakly similar to GP|3128176|gb|AAC16080.1| unknown prot... 42 8e-05
TC79217 weakly similar to GP|21553541|gb|AAM62634.1 lipid transf... 41 2e-04
TC79755 weakly similar to PIR|T04972|T04972 hypothetical protein... 39 9e-04
TC86318 similar to GP|15451110|gb|AAK96826.1 Unknown protein {Ar... 37 0.002
TC76731 similar to GP|13937151|gb|AAK50069.1 AT5g57180/MUL3_13 {... 37 0.003
TC86319 similar to GP|15451110|gb|AAK96826.1 Unknown protein {Ar... 37 0.003
BE319937 similar to PIR|T09546|T095 extensin like protein - blac... 35 0.007
TC76828 similar to GP|17154773|gb|AAL35979.1 extensin-like prote... 35 0.013
TC76827 similar to GP|17154773|gb|AAL35979.1 extensin-like prote... 35 0.013
TC88967 weakly similar to GP|18477856|emb|CAC86258. lipid transf... 33 0.028
TC76916 MtN4 33 0.037
>TC87425 weakly similar to GP|20161478|dbj|BAB90402. contains ESTs
C74501(E31745) AU094804(E31745)~unknown protein, partial
(24%)
Length = 929
Score = 259 bits (663), Expect = 2e-70
Identities = 128/128 (100%), Positives = 128/128 (100%)
Frame = +1
Query: 4 TTLVVAIVSLIFASLSFHGDGVSAAALAQSPAPETAVLAPSPADDGCLMALTNMSDCLTF 63
TTLVVAIVSLIFASLSFHGDGVSAAALAQSPAPETAVLAPSPADDGCLMALTNMSDCLTF
Sbjct: 70 TTLVVAIVSLIFASLSFHGDGVSAAALAQSPAPETAVLAPSPADDGCLMALTNMSDCLTF 249
Query: 64 VEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSNTADSFGIKINVNKALKLPTICGVT 123
VEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSNTADSFGIKINVNKALKLPTICGVT
Sbjct: 250 VEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSNTADSFGIKINVNKALKLPTICGVT 429
Query: 124 TPPVSACS 131
TPPVSACS
Sbjct: 430 TPPVSACS 453
>BG454079 similar to GP|10176956|dbj contains similarity to nonspecific
lipid-transfer protein~gene_id:MHJ24.6 {Arabidopsis
thaliana}, partial (50%)
Length = 418
Score = 81.3 bits (199), Expect = 1e-16
Identities = 41/97 (42%), Positives = 56/97 (57%)
Frame = +1
Query: 42 APSPADDGCLMALTNMSDCLTFVEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSNTA 101
AP+P+ D C + M+DCL+FV +GS TKP+ CC L ++ P CLC+ S+
Sbjct: 100 APAPSVD-CTNLVLTMADCLSFVTNGSTTTKPEGTCCSGLKSVLKTAPSCLCEAFKSSA- 273
Query: 102 DSFGIKINVNKALKLPTICGVTTPPVSACSGKFFMSL 138
FG+ +NV KA LP C V+ P + C KFF SL
Sbjct: 274 -QFGVVLNVTKATSLPAACKVSAPSATKCGCKFFSSL 381
>TC79365 weakly similar to GP|15010698|gb|AAK74008.1 AT3g22600/F16J14_17
{Arabidopsis thaliana}, partial (51%)
Length = 783
Score = 78.2 bits (191), Expect = 1e-15
Identities = 37/87 (42%), Positives = 49/87 (55%)
Frame = +2
Query: 46 ADDGCLMALTNMSDCLTFVEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSNTADSFG 105
A GC +T++S CL ++ S + P CC +L+ ++ +P CLC LL SFG
Sbjct: 122 AQSGCTSTITSLSPCLNYIMGSS--SNPSSSCCSQLSSVVQSSPQCLCSLLNGG-GSSFG 292
Query: 106 IKINVNKALKLPTICGVTTPPVSACSG 132
I IN AL LP+ C V TPPVS C G
Sbjct: 293 ITINQTLALSLPSACKVQTPPVSQCKG 373
>TC78493 weakly similar to GP|15010698|gb|AAK74008.1 AT3g22600/F16J14_17
{Arabidopsis thaliana}, partial (57%)
Length = 797
Score = 76.3 bits (186), Expect = 4e-15
Identities = 37/89 (41%), Positives = 49/89 (54%)
Frame = +2
Query: 42 APSPADDGCLMALTNMSDCLTFVEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSNTA 101
A + A C L ++S CL ++ S + P GCC LA ++ P+CLC++LG A
Sbjct: 128 AGATAQSSCTNVLVSLSPCLNYITGNS--STPSSGCCSNLASVVSSQPLCLCQVLGGG-A 298
Query: 102 DSFGIKINVNKALKLPTICGVTTPPVSAC 130
S GI IN +AL LP C V TPP S C
Sbjct: 299 SSLGISINQTQALALPGACKVQTPPTSQC 385
>TC87440 similar to GP|17979494|gb|AAL50083.1 AT5g64080/MHJ24_6 {Arabidopsis
thaliana}, partial (48%)
Length = 818
Score = 73.2 bits (178), Expect = 3e-14
Identities = 36/89 (40%), Positives = 51/89 (56%)
Frame = +1
Query: 42 APSPADDGCLMALTNMSDCLTFVEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSNTA 101
AP+P+ D C + M+DCL+FV +GS TKP+ CC L ++ P CLC+ S+
Sbjct: 112 APAPSVD-CTNLVLTMADCLSFVTNGSTTTKPEGTCCSGLKSVLKTAPSCLCEAFKSSA- 285
Query: 102 DSFGIKINVNKALKLPTICGVTTPPVSAC 130
FG+ +NV KA LP C V+ P + C
Sbjct: 286 -QFGVVLNVTKATSLPAACKVSAPSATKC 369
>TC90904 similar to GP|3128176|gb|AAC16080.1| unknown protein {Arabidopsis
thaliana}, partial (28%)
Length = 672
Score = 55.8 bits (133), Expect = 5e-09
Identities = 31/72 (43%), Positives = 38/72 (52%), Gaps = 1/72 (1%)
Frame = +3
Query: 50 CLMALTNMSDCLTFVEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGS-NTADSFGIKI 108
C LT ++ CL +VE K PD CC L L+ N CLC ++ N D GI I
Sbjct: 159 CTAQLTGLASCLPYVEGEGKTPAPD--CCDGLKTLLKTNKKCLCVIIKDRNDPDLGGIVI 332
Query: 109 NVNKALKLPTIC 120
NV AL LPT+C
Sbjct: 333 NVTLALNLPTVC 368
>BF520250 weakly similar to GP|3128176|gb| unknown protein {Arabidopsis
thaliana}, partial (69%)
Length = 541
Score = 47.4 bits (111), Expect = 2e-06
Identities = 26/75 (34%), Positives = 37/75 (48%)
Frame = +3
Query: 50 CLMALTNMSDCLTFVEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSNTADSFGIKIN 109
C L ++ CL FV + +K P CC + + D + CLC L+ + + G+ IN
Sbjct: 126 CTNKLLTLAGCLPFVTNQAK--SPTIDCCTGVKEVADKSKRCLCILIKDHDDPNLGLTIN 299
Query: 110 VNKALKLPTICGVTT 124
V ALKLP C T
Sbjct: 300 VTLALKLPNDCNSPT 344
>TC81134 weakly similar to GP|14334664|gb|AAK59510.1 unknown protein
{Arabidopsis thaliana}, partial (42%)
Length = 900
Score = 42.7 bits (99), Expect = 5e-05
Identities = 29/95 (30%), Positives = 46/95 (47%), Gaps = 5/95 (5%)
Frame = +3
Query: 38 TAVLAPSPAD--DGCLMAL--TNMSDCLTFVEDGS-KLTKPDKGCCPELAGLIDGNPICL 92
TA+ AP+ A C M++ + +S CL+F+ + S T P CC + L G+ C+
Sbjct: 108 TAMAAPAYAQITTPCNMSMISSTISPCLSFLTNSSGNGTSPTADCCNAIKTLTSGSKDCM 287
Query: 93 CKLLGSNTADSFGIKINVNKALKLPTICGVTTPPV 127
C + N F + IN A+ LP C + P+
Sbjct: 288 CLIATGNV--PFALPINRTLAISLPRACNLPGVPL 386
>AJ548147 similar to GP|15010698|gb| AT3g22600/F16J14_17 {Arabidopsis
thaliana}, partial (26%)
Length = 390
Score = 42.7 bits (99), Expect = 5e-05
Identities = 20/42 (47%), Positives = 24/42 (56%)
Frame = -1
Query: 89 PICLCKLLGSNTADSFGIKINVNKALKLPTICGVTTPPVSAC 130
P CLC++L S GIK+N +AL P C V TPP S C
Sbjct: 384 PQCLCQVLDGG-GSSLGIKVNQTQALAWPCACNVQTPPTSQC 262
>TC78186 weakly similar to GP|3128176|gb|AAC16080.1| unknown protein
{Arabidopsis thaliana}, partial (32%)
Length = 766
Score = 42.0 bits (97), Expect = 8e-05
Identities = 21/81 (25%), Positives = 36/81 (43%)
Frame = +2
Query: 50 CLMALTNMSDCLTFVEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSNTADSFGIKIN 109
C L ++ CL +V G P CC L +++ C+C L+ + G +N
Sbjct: 95 CADKLVTLASCLPYV--GGSANTPTIDCCTNLKQVLNNTKKCICILIKDSNDPKLGFPMN 268
Query: 110 VNKALKLPTICGVTTPPVSAC 130
A++LP C + + +S C
Sbjct: 269 ATLAVQLPNACHIPS-NISEC 328
>TC79217 weakly similar to GP|21553541|gb|AAM62634.1 lipid transfer protein
putative {Arabidopsis thaliana}, partial (43%)
Length = 992
Score = 40.8 bits (94), Expect = 2e-04
Identities = 27/86 (31%), Positives = 39/86 (44%), Gaps = 5/86 (5%)
Frame = +1
Query: 50 CLMALTNMSDCLTFVEDGSKLTKPDKGCCPELAGLIDGNPICLCKLL-----GSNTADSF 104
C + + CL F K P K CC + +P CLC ++ GS + S
Sbjct: 271 CGSVVQKVIPCLDFAT--GKAPTPKKECCDAANSIKATDPECLCYIIQQTHKGSPESKSM 444
Query: 105 GIKINVNKALKLPTICGVTTPPVSAC 130
GI+ +K L+LPT+C V +S C
Sbjct: 445 GIQ--EDKLLQLPTVCHVNGANISDC 516
>TC79755 weakly similar to PIR|T04972|T04972 hypothetical protein T16L1.40 -
Arabidopsis thaliana, partial (33%)
Length = 707
Score = 38.5 bits (88), Expect = 9e-04
Identities = 27/82 (32%), Positives = 39/82 (46%)
Frame = +3
Query: 49 GCLMALTNMSDCLTFVEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSNTADSFGIKI 108
G L AL +S+C FV+ P GCC A + + C CKL+ + A+ F +
Sbjct: 141 GSLPAL--ISECSKFVQKSGPKIAPSPGCC---AAIRSFDVPCACKLI-TKEAEKF---V 293
Query: 109 NVNKALKLPTICGVTTPPVSAC 130
+V KA+ + CGV P C
Sbjct: 294 SVPKAISVARSCGVKLPAGMQC 359
>TC86318 similar to GP|15451110|gb|AAK96826.1 Unknown protein {Arabidopsis
thaliana}, partial (79%)
Length = 618
Score = 37.4 bits (85), Expect = 0.002
Identities = 33/131 (25%), Positives = 52/131 (39%), Gaps = 1/131 (0%)
Frame = +2
Query: 5 TLVVAIVSLIFASLSFHGDGVSAAALAQSPAPETAV-LAPSPADDGCLMALTNMSDCLTF 63
T + +V + AS F + +A ++P A L+P CL A+ N+
Sbjct: 74 TFLATVVMFLLASSVFILESEAAGECGRTPIGSAAASLSP------CLGAVRNVR----- 220
Query: 64 VEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSNTADSFGIKINVNKALKLPTICGVT 123
K CC + L+ +P CLC +L S A KIN A+ +P C +
Sbjct: 221 -------AKVPPVCCARVGALLRTSPRCLCSVLLSPLAKQ--AKINPAIAITVPKRCNIR 373
Query: 124 TPPVSACSGKF 134
P G++
Sbjct: 374 NRPAGKKCGRY 406
>TC76731 similar to GP|13937151|gb|AAK50069.1 AT5g57180/MUL3_13 {Arabidopsis
thaliana}, partial (10%)
Length = 1235
Score = 37.0 bits (84), Expect = 0.003
Identities = 23/71 (32%), Positives = 31/71 (43%), Gaps = 1/71 (1%)
Frame = +3
Query: 55 TNMSDCLTFVEDGSKLTKPDKGCCPELAGLIDGNPICLCKLL-GSNTADSFGIKINVNKA 113
TN+ C ++ TKP CC + ++ CLC L +F IN +A
Sbjct: 597 TNLIPCADYLNS----TKPPSSCCDPIKKTVETELTCLCNLFYAPGLLATF--NINTTQA 758
Query: 114 LKLPTICGVTT 124
L L CGVTT
Sbjct: 759 LALSRNCGVTT 791
>TC86319 similar to GP|15451110|gb|AAK96826.1 Unknown protein {Arabidopsis
thaliana}, partial (79%)
Length = 760
Score = 36.6 bits (83), Expect = 0.003
Identities = 24/82 (29%), Positives = 36/82 (43%)
Frame = +3
Query: 53 ALTNMSDCLTFVEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSNTADSFGIKINVNK 112
A ++S CL V + P CC + L+ +P CLC +L S A KIN
Sbjct: 162 AAASLSPCLGAVRNVRAKVPPV--CCARVGALLRTSPRCLCSVLLSPLAKQ--AKINPAI 329
Query: 113 ALKLPTICGVTTPPVSACSGKF 134
A+ +P C + P G++
Sbjct: 330 AITVPKRCNIRNRPAGKKCGRY 395
>BE319937 similar to PIR|T09546|T095 extensin like protein - black poplar,
partial (67%)
Length = 423
Score = 35.4 bits (80), Expect = 0.007
Identities = 31/132 (23%), Positives = 57/132 (42%), Gaps = 7/132 (5%)
Frame = +3
Query: 2 ATTTLVVAIVSLIFASLSFHGDGVSAAALAQSPAPETAVLA---PSPADDGCLMALTNMS 58
++T ++ +I++++F +++ + P P PSP+ C +
Sbjct: 21 SSTLVLFSIINMLFFAMA------NGCFFCPKPNPNPNPFPYPNPSPSTKSCPRDALKLG 182
Query: 59 DCLTFVED--GSKL-TKPDKGCCPELAGLIDGN-PICLCKLLGSNTADSFGIKINVNKAL 114
C + G+ + + P+ CC L GL+D +CLC + +N GI IN+ +L
Sbjct: 183 VCANLLNGPIGAVIGSPPEHPCCSILEGLVDLEVAVCLCTAIKANI---LGIDINIPISL 353
Query: 115 KLPTICGVTTPP 126
L TPP
Sbjct: 354 SLILNACEKTPP 389
>TC76828 similar to GP|17154773|gb|AAL35979.1 extensin-like protein {Cucumis
sativus}, partial (60%)
Length = 739
Score = 34.7 bits (78), Expect = 0.013
Identities = 28/91 (30%), Positives = 37/91 (39%), Gaps = 4/91 (4%)
Frame = +1
Query: 45 PADDGCLMALTNMSDCLTFVEDGSKLTKPD---KGCCPELAGLIDGNPI-CLCKLLGSNT 100
PA D C + + C+ + + D CCP L GL + CLC L
Sbjct: 427 PAKDTCPIDTLKLGACVDLLGGLVHIGLGDPVVNKCCPVLQGLAEIEAAACLCTTLKLKL 606
Query: 101 ADSFGIKINVNKALKLPTICGVTTPPVSACS 131
+ + I V AL+L CG T PP CS
Sbjct: 607 LN---LNIYVPLALQLLLTCGKTPPPGYTCS 690
>TC76827 similar to GP|17154773|gb|AAL35979.1 extensin-like protein {Cucumis
sativus}, partial (60%)
Length = 1093
Score = 34.7 bits (78), Expect = 0.013
Identities = 28/91 (30%), Positives = 37/91 (39%), Gaps = 4/91 (4%)
Frame = +3
Query: 45 PADDGCLMALTNMSDCLTFVEDGSKLTKPD---KGCCPELAGLIDGNPI-CLCKLLGSNT 100
PA D C + + C+ + + D CCP L GL + CLC L
Sbjct: 534 PAKDTCPIDTLKLGACVDLLGGLVHIGLGDPVVNKCCPVLQGLAEIEAAACLCTTLKLKL 713
Query: 101 ADSFGIKINVNKALKLPTICGVTTPPVSACS 131
+ + I V AL+L CG T PP CS
Sbjct: 714 LN---LNIYVPLALQLLLTCGKTPPPGYTCS 797
>TC88967 weakly similar to GP|18477856|emb|CAC86258. lipid transfer protein
{Fragaria x ananassa}, partial (48%)
Length = 743
Score = 33.5 bits (75), Expect = 0.028
Identities = 32/132 (24%), Positives = 50/132 (37%), Gaps = 7/132 (5%)
Frame = +1
Query: 1 MATTTLVVAIVSLIFASLSFHGDGVSAAALAQSPAPETAVLAPSPADDGCLMALTNMSDC 60
MAT + +AIV L+ P E AV + P +T++ C
Sbjct: 61 MATRLVCLAIVCLV----------------TFGPKAEAAVTSCGPV-------VTSLYPC 171
Query: 61 LTFVEDGSKLTKPDKGCCPELAGLI-------DGNPICLCKLLGSNTADSFGIKINVNKA 113
++++ +G T P CC + L D +C C + + +N+N A
Sbjct: 172 VSYIMNGGN-TVPAAQCCNGIRNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLA 348
Query: 114 LKLPTICGVTTP 125
LP CGV P
Sbjct: 349 AGLPRKCGVNIP 384
>TC76916 MtN4
Length = 1850
Score = 33.1 bits (74), Expect = 0.037
Identities = 26/101 (25%), Positives = 38/101 (36%), Gaps = 4/101 (3%)
Frame = +1
Query: 34 PAPETAVLAPSPADDGCLMALTNMSDCLTFVEDGSKLT---KPDKGCCPELAGLID-GNP 89
P P P PA C + + C+ + + + CCP L GL+D
Sbjct: 1135 PPPPLVPYPPPPAQQTCSIDALKLGACVDVLGGLIHIGIGGSAKQTCCPLLQGLVDLDAA 1314
Query: 90 ICLCKLLGSNTADSFGIKINVNKALKLPTICGVTTPPVSAC 130
ICLC + I + + AL++ CG T P C
Sbjct: 1315 ICLCTTI---RLKLLNINLVIPLALQVLIDCGKTPPEGFKC 1428
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.135 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,698,846
Number of Sequences: 36976
Number of extensions: 68050
Number of successful extensions: 390
Number of sequences better than 10.0: 60
Number of HSP's better than 10.0 without gapping: 384
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 386
length of query: 139
length of database: 9,014,727
effective HSP length: 86
effective length of query: 53
effective length of database: 5,834,791
effective search space: 309243923
effective search space used: 309243923
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 53 (25.0 bits)
Medicago: description of AC146743.12


