

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146723.15 + phase: 0
(345 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
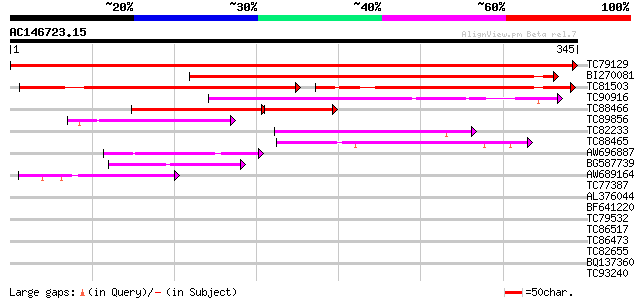
Score E
Sequences producing significant alignments: (bits) Value
TC79129 similar to PIR|A96725|A96725 hypothetical protein F20P5.... 680 0.0
BI270081 similar to GP|6448480|emb cyclin D1 {Antirrhinum majus}... 344 2e-95
TC81503 similar to GP|6448480|emb|CAB61221.1 cyclin D1 {Antirrhi... 176 1e-44
TC90916 similar to PIR|T09961|T09961 cyclin D-like protein - red... 118 4e-27
TC88466 similar to GP|6434199|emb|CAB60837.1 CycD3;2 {Lycopersic... 85 3e-24
TC89856 homologue to GP|4583990|emb|CAB40540.1 cyclin D3 {Medica... 65 5e-11
TC82233 homologue to PIR|T09598|T09598 cyclin 4 D-type - alfalf... 62 3e-10
TC88465 similar to GP|6434201|emb|CAB60838.1 CycD3;3 {Lycopersic... 60 2e-09
AW696887 similar to PIR|T07669|T07 cyclin a1-type mitosis-speci... 58 5e-09
BG587739 GP|18147003|db cyclin D2 {Sus scrofa}, partial (3%) 57 1e-08
AW689164 weakly similar to GP|19070617|gb| D-type cyclin {Zea ma... 54 9e-08
TC77387 similar to GP|15983396|gb|AAL11566.1 At1g51440/F5D21_19 ... 32 0.36
AL376044 similar to GP|18656155|db ryanodine receptor {Hemicentr... 31 0.61
BF641220 weakly similar to PIR|G86203|G86 probable N-arginine di... 31 0.80
TC79532 weakly similar to GP|8164030|gb|AAF73960.1| SGS3 {Arabid... 31 0.80
TC86517 homologue to PIR|T09640|T09640 protein phosphatase 2C - ... 30 1.0
TC86473 similar to PIR|E84828|E84828 probable WD-40 repeat prote... 30 1.0
TC82655 similar to PIR|T49930|T49930 hypothetical protein F17I14... 30 1.0
BQ137360 30 1.0
TC93240 weakly similar to GP|14335170|gb|AAK59865.1 AT5g51070/K3... 30 1.8
>TC79129 similar to PIR|A96725|A96725 hypothetical protein F20P5.7
[imported] - Arabidopsis thaliana, partial (64%)
Length = 1328
Score = 680 bits (1755), Expect = 0.0
Identities = 343/345 (99%), Positives = 344/345 (99%)
Frame = +2
Query: 1 MPLSSTSDCELLCGEDSSEVLTGDLPECSSDLDSSSSSQLPSSSLFAEEEEESIAVFIEH 60
MPLSSTSDCELLCGEDSSEVLTGDLPECSSDLDSSSSSQLPSSSLFAEEEEESIAVFIEH
Sbjct: 113 MPLSSTSDCELLCGEDSSEVLTGDLPECSSDLDSSSSSQLPSSSLFAEEEEESIAVFIEH 292
Query: 61 EFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRP 120
EFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRP
Sbjct: 293 EFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRP 472
Query: 121 LPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILD 180
LPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILD
Sbjct: 473 LPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILD 652
Query: 181 WRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFLTYRPSCIAAAAIL 240
WRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFLTYRPSCIAAAAIL
Sbjct: 653 WRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFLTYRPSCIAAAAIL 832
Query: 241 SAANEIPNWSFVNPEHAESWCEGLSKEKIIGCYELIQEIVSSNNQRNAPKVLPQLRVTAR 300
SAANEIPNWSFVNPEHAESWCEGLSKEKIIGCYELIQEIVSSNNQRNAPKVLPQLRVTAR
Sbjct: 833 SAANEIPNWSFVNPEHAESWCEGLSKEKIIGCYELIQEIVSSNNQRNAPKVLPQLRVTAR 1012
Query: 301 TRRWSTVSSSSSSPSSSSSPSFSLSYKKRKLNSCFWVDVDKGNSE 345
TRRWSTVSSSSSSPSSSSSPSFSLSYKK+KLNS FWVDVDKGNSE
Sbjct: 1013TRRWSTVSSSSSSPSSSSSPSFSLSYKKKKLNSWFWVDVDKGNSE 1147
>BI270081 similar to GP|6448480|emb cyclin D1 {Antirrhinum majus}, partial
(50%)
Length = 663
Score = 344 bits (883), Expect = 2e-95
Identities = 174/225 (77%), Positives = 193/225 (85%)
Frame = +2
Query: 110 NYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTIL 169
NYMDRFL+SR LP++NGWPLQLLSVACLSLAAKMEE LVPSLLD Q+EG KY+F+P TI
Sbjct: 2 NYMDRFLNSRRLPQTNGWPLQLLSVACLSLAAKMEETLVPSLLDLQVEGVKYMFEPITIR 181
Query: 170 RMELLVLTILDWRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFLTY 229
RMELLVL++LDWRLRS+TP SFLSFFACKLDST TFT F+ISRAT+IILS IQ+AS L Y
Sbjct: 182 RMELLVLSVLDWRLRSVTPFSFLSFFACKLDSTSTFTGFLISRATQIILSKIQEASILAY 361
Query: 230 RPSCIAAAAILSAANEIPNWSFVNPEHAESWCEGLSKEKIIGCYELIQEIVSSNNQRNAP 289
PSCIAAAAIL AANEIPNWS V PEHAESWCEGL KEKIIGCY+L+QE+V NNQR P
Sbjct: 362 WPSCIAAAAILYAANEIPNWSLVEPEHAESWCEGLRKEKIIGCYQLMQELVIDNNQRKPP 541
Query: 290 KVLPQLRVTARTRRWSTVSSSSSSPSSSSSPSFSLSYKKRKLNSC 334
KVLPQ+RVT + S VSSSSSSPSSSSS SYK+RKLN+C
Sbjct: 542 KVLPQMRVTIQPLMRSCVSSSSSSPSSSSS-----SYKRRKLNNC 661
>TC81503 similar to GP|6448480|emb|CAB61221.1 cyclin D1 {Antirrhinum majus},
partial (36%)
Length = 1000
Score = 176 bits (446), Expect = 1e-44
Identities = 97/158 (61%), Positives = 111/158 (69%)
Frame = +2
Query: 187 TPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFLTYRPSCIAAAAILSAANEI 246
T +S L F+CK F+ F I + +AS L Y PSCIAAAAIL AANEI
Sbjct: 524 TSISCLLVFSCK--DGRNFSPFSIGPS---------EASILAYWPSCIAAAAILYAANEI 670
Query: 247 PNWSFVNPEHAESWCEGLSKEKIIGCYELIQEIVSSNNQRNAPKVLPQLRVTARTRRWST 306
PNWS V PEHAESWCEGL KEKIIGCY+L+QE+V NNQR PKVLPQ+RVT + S
Sbjct: 671 PNWSLVEPEHAESWCEGLRKEKIIGCYQLMQELVIDNNQRKPPKVLPQMRVTIQPLMRSC 850
Query: 307 VSSSSSSPSSSSSPSFSLSYKKRKLNSCFWVDVDKGNS 344
VSSSSSSPSSSSS SYK+RKLN+C WVD DKG++
Sbjct: 851 VSSSSSSPSSSSS-----SYKRRKLNNCLWVDDDKGSN 949
Score = 169 bits (427), Expect = 2e-42
Identities = 93/172 (54%), Positives = 117/172 (67%), Gaps = 1/172 (0%)
Frame = +3
Query: 7 SDCELLCGEDSSEVLTGDLP-ECSSDLDSSSSSQLPSSSLFAEEEEESIAVFIEHEFKFV 65
+D +LLCGE++S +L+ D P E SD +S + E+E IA IE + KFV
Sbjct: 183 TDSDLLCGEETSSILSSDSPTESFSDGES-----------YPPPEDEFIAGLIEDQGKFV 329
Query: 66 PGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPESN 125
GFDY + +S S +S R+E+I WILKV YYGFQP+TAYL+VNYMDRFL+SR LP++N
Sbjct: 330 IGFDYFVKMKSSSFDSDARDESIRWILKVQGYYGFQPVTAYLAVNYMDRFLNSRRLPQTN 509
Query: 126 GWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLT 177
GWPLQLLSVACLSLAAKMEE LVPSLLD Q +I + R+ + LT
Sbjct: 510 GWPLQLLSVACLSLAAKMEETLVPSLLDLQRPAFLHIGPHALLPRLYSMQLT 665
>TC90916 similar to PIR|T09961|T09961 cyclin D-like protein - red goosefoot,
partial (31%)
Length = 886
Score = 118 bits (295), Expect = 4e-27
Identities = 79/221 (35%), Positives = 117/221 (52%), Gaps = 6/221 (2%)
Frame = +3
Query: 122 PESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILDW 181
P+ W +QLL+VACLSLAAK+EE VP LD QI +K++F+ +TI RMELLVL+ L W
Sbjct: 15 PKGRAWTMQLLAVACLSLAAKVEETAVPQPLDLQIGESKFVFEAKTIQRMELLVLSTLKW 194
Query: 182 RLRSITPLSFLSFFACKL-DSTGTFTHFIISRATEIILSNIQDASFLTYRPSCIAAAAIL 240
R+++ITP SF+ F K+ D + ISR+T++I S I+ FL ++PS IAAA
Sbjct: 195 RMQAITPFSFIECFLSKIKDDDKSSLSSSISRSTQLISSTIKGLDFLEFKPSEIAAAVAT 374
Query: 241 SAANEIPNWSFVNPEHAESWCEGLSKEKIIGCYELIQEIVSSNNQRNAPKVLPQLRVTAR 300
E + + + + + + K +++ C +QE +S N+
Sbjct: 375 CVVGE--TQAIDSSKSISTLIQYVEKGRLLKCVGKVQE-MSLNSVFTGKD---------- 515
Query: 301 TRRWSTVSSSSSSPSSSSSP-----SFSLSYKKRKLNSCFW 336
SS+SS PS SP + SYK N+ +W
Sbjct: 516 -------SSASSVPSVPQSPMGVLDTLCFSYKSDDTNAGWW 617
>TC88466 similar to GP|6434199|emb|CAB60837.1 CycD3;2 {Lycopersicon
esculentum}, partial (39%)
Length = 692
Score = 84.7 bits (208), Expect(2) = 3e-24
Identities = 43/82 (52%), Positives = 54/82 (65%)
Frame = +2
Query: 75 QSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPESNGWPLQLLSV 134
Q+ + + R E+I WILKV+ +Y F LT+ L+VNY+DRFL S W QL +V
Sbjct: 170 QTNPVLETARRESIEWILKVNAHYSFSALTSVLAVNYLDRFLFSFRFQNEKPWMTQLAAV 349
Query: 135 ACLSLAAKMEEPLVPSLLDFQI 156
ACLSLAAKMEE VP LLD Q+
Sbjct: 350 ACLSLAAKMEETHVPLLLDLQV 415
Score = 44.7 bits (104), Expect(2) = 3e-24
Identities = 17/44 (38%), Positives = 31/44 (69%)
Frame = +1
Query: 156 IEGAKYIFQPRTILRMELLVLTILDWRLRSITPLSFLSFFACKL 199
+E ++Y+F+ +TI +ME+L+L+ L W++ TPLSF+ +L
Sbjct: 487 VEESRYLFEAKTIKKMEILILSTLGWKMNPATPLSFIDXIIRRL 618
>TC89856 homologue to GP|4583990|emb|CAB40540.1 cyclin D3 {Medicago sativa},
partial (41%)
Length = 689
Score = 64.7 bits (156), Expect = 5e-11
Identities = 37/106 (34%), Positives = 61/106 (56%), Gaps = 4/106 (3%)
Frame = +1
Query: 36 SSSQLP----SSSLFAEEEEESIAVFIEHEFKFVPGFDYVSRFQSRSLESSTREEAIAWI 91
S+S LP +LF ++EE + +F + + + +D + + + R EA+ W+
Sbjct: 373 STSLLPLLLLEQNLFNKDEELN-TLFSKEKIQQETYYDDLKNVMNFDSLTQPRREAVEWM 549
Query: 92 LKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPESNGWPLQLLSVACL 137
LKV+ +YGF LTA L+VNY+DRFL S + W +QL++V C+
Sbjct: 550 LKVNAHYGFSALTATLAVNYLDRFLLSFHFQKEKPWMIQLVAVTCI 687
>TC82233 homologue to PIR|T09598|T09598 cyclin 4 D-type - alfalfa, partial
(51%)
Length = 1011
Score = 62.0 bits (149), Expect = 3e-10
Identities = 37/125 (29%), Positives = 63/125 (49%), Gaps = 2/125 (1%)
Frame = +1
Query: 162 IFQPRTILRMELLVLTILDWRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNI 221
+F+ +TI RMELL+L+ L W++ +T SFL +L + R ++LS +
Sbjct: 1 VFEAKTIQRMELLILSTLKWKMHPVTTHSFLDHIIRRLGLKTNLHWEFLRRCENLLLSVL 180
Query: 222 QDASFLTYRPSCIAAAAILSAANEIPNWSFVNPEHAESWCEGL--SKEKIIGCYELIQEI 279
D+ F+ PS +A A +L ++I + ++ L SKEK+ CY I +
Sbjct: 181 LDSRFVGCVPSVLATATMLHVIDQIEQSDDNDVDYKNQLLNVLKISKEKVDECYNAILHL 360
Query: 280 VSSNN 284
++NN
Sbjct: 361 TNTNN 375
>TC88465 similar to GP|6434201|emb|CAB60838.1 CycD3;3 {Lycopersicon
esculentum}, partial (30%)
Length = 637
Score = 59.7 bits (143), Expect = 2e-09
Identities = 47/176 (26%), Positives = 87/176 (48%), Gaps = 20/176 (11%)
Frame = +3
Query: 163 FQPRTILRMELLVLTILDWRLRSITPLSFLSFFACKLDSTGTFTHFI----ISRATEIIL 218
F+ +TI +ME+L+L+ L W++ TPLSF+ F +L G H I + R ++L
Sbjct: 9 FEAKTIKKMEILILSTLGWKMNPATPLSFIDFIIRRL---GLKDHLICWEFLKRCEGVLL 179
Query: 219 SNIQ-DASFLTYRPSCIAAAAILSAANEIPNWSFVNPEHAESWCEGLSKEKIIGCYELIQ 277
S I+ D+ F++Y PS +A A ++ N + + G++K+K+ C +L+
Sbjct: 180 SVIRSDSKFMSYLPSVLATATMVHVFNSVEPSLGDEYQTQLLGILGINKDKVDECGKLLL 359
Query: 278 EIVSSNNQRN------------APKVLPQLRVTARTRR--WSTVSSS-SSSPSSSS 318
++ S + N +PK + ++ + W+ +++S SSSP S
Sbjct: 360 KLWSGYEEGNECNKRKFGSIPSSPKGVMEMSFSCDNSNDSWAIIAASVSSSPEPLS 527
>AW696887 similar to PIR|T07669|T07 cyclin a1-type mitosis-specific -
soybean, partial (33%)
Length = 648
Score = 58.2 bits (139), Expect = 5e-09
Identities = 35/97 (36%), Positives = 54/97 (55%)
Frame = +3
Query: 58 IEHEFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLD 117
+E + K P Y+ Q R + S+ R + W+++V + Y P T +LSV+Y+DRFL
Sbjct: 366 MEMQKKRRPMVGYLENVQ-RGISSNMRGTLVDWLVEVADEYKLLPETLHLSVSYIDRFLS 542
Query: 118 SRPLPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDF 154
P+ S LQLL V+ + +A+K EE P +DF
Sbjct: 543 IEPVSRSK---LQLLGVSSMLIASKYEEITPPKAVDF 644
>BG587739 GP|18147003|db cyclin D2 {Sus scrofa}, partial (3%)
Length = 519
Score = 56.6 bits (135), Expect = 1e-08
Identities = 28/83 (33%), Positives = 46/83 (54%)
Frame = +3
Query: 61 EFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRP 120
E +P +Y +S ++S R + I+ I ++ F P YL++NY+DRFL ++
Sbjct: 213 ESDHIPPLNYFQNLKSNEFDASVRTDFISLISQLS--CNFDPFVTYLAINYLDRFLANQG 386
Query: 121 LPESNGWPLQLLSVACLSLAAKM 143
+ + W +LL+V C SLA KM
Sbjct: 387 ILQPKPWANKLLAVTCFSLAVKM 455
>AW689164 weakly similar to GP|19070617|gb| D-type cyclin {Zea mays}, partial
(27%)
Length = 463
Score = 53.9 bits (128), Expect = 9e-08
Identities = 36/108 (33%), Positives = 53/108 (48%), Gaps = 10/108 (9%)
Frame = +1
Query: 6 TSDCELLCGEDSS----EVLTGDLPECSS------DLDSSSSSQLPSSSLFAEEEEESIA 55
+++ LLC E++S +V+ D S +LD+ S S F + EE +
Sbjct: 148 SAESNLLCSENNSTCFDDVVVDDSGISPSWDHTNVNLDNVGSD---SFLCFVAQSEEIVK 318
Query: 56 VFIEHEFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPL 103
V +E E +P DY+ R + L+ S EA+ WI K H YYGF PL
Sbjct: 319 VMVEKEKDHLPREDYLIRLRGGDLDLSVTREALDWIWKAHAYYGFGPL 462
>TC77387 similar to GP|15983396|gb|AAL11566.1 At1g51440/F5D21_19
{Arabidopsis thaliana}, partial (75%)
Length = 1793
Score = 32.0 bits (71), Expect = 0.36
Identities = 18/33 (54%), Positives = 20/33 (60%)
Frame = -2
Query: 309 SSSSSPSSSSSPSFSLSYKKRKLNSCFWVDVDK 341
SSSSS SSSSS S S S +SC W VD+
Sbjct: 397 SSSSSSSSSSSSSSSSSSSSSSSSSCSWFIVDE 299
>AL376044 similar to GP|18656155|db ryanodine receptor {Hemicentrotus
pulcherrimus}, partial (0%)
Length = 525
Score = 31.2 bits (69), Expect = 0.61
Identities = 21/45 (46%), Positives = 27/45 (59%)
Frame = -1
Query: 289 PKVLPQLRVTARTRRWSTVSSSSSSPSSSSSPSFSLSYKKRKLNS 333
P+ P L + A ++SSSSSSPSSSSS S S S R ++S
Sbjct: 195 PRAFPTLGI*AGFLAGCSLSSSSSSPSSSSSSS-SPSRSSRSISS 64
>BF641220 weakly similar to PIR|G86203|G86 probable N-arginine dibasic
convertase [imported] - Arabidopsis thaliana, partial
(5%)
Length = 634
Score = 30.8 bits (68), Expect = 0.80
Identities = 19/33 (57%), Positives = 20/33 (60%)
Frame = -2
Query: 293 PQLRVTARTRRWSTVSSSSSSPSSSSSPSFSLS 325
P L T + S SSSSSSPSSSSS S S S
Sbjct: 486 PSLPXTXXSSSSSXSSSSSSSPSSSSSSSSSSS 388
>TC79532 weakly similar to GP|8164030|gb|AAF73960.1| SGS3 {Arabidopsis
thaliana}, partial (47%)
Length = 1558
Score = 30.8 bits (68), Expect = 0.80
Identities = 18/31 (58%), Positives = 22/31 (70%)
Frame = -3
Query: 295 LRVTARTRRWSTVSSSSSSPSSSSSPSFSLS 325
+R ++ + R S SSSSSS SSSSSPSF S
Sbjct: 731 IRSSSVSSRSSQSSSSSSSSSSSSSPSFFCS 639
>TC86517 homologue to PIR|T09640|T09640 protein phosphatase 2C - alfalfa,
complete
Length = 1616
Score = 30.4 bits (67), Expect = 1.0
Identities = 18/39 (46%), Positives = 22/39 (56%)
Frame = +2
Query: 305 STVSSSSSSPSSSSSPSFSLSYKKRKLNSCFWVDVDKGN 343
+T SSSSSSPS SSPS + KL F + D G+
Sbjct: 254 TTSSSSSSSPSPCSSPSSPFCLRLPKLPLVFTSNKDSGS 370
>TC86473 similar to PIR|E84828|E84828 probable WD-40 repeat protein
[imported] - Arabidopsis thaliana, partial (68%)
Length = 2654
Score = 30.4 bits (67), Expect = 1.0
Identities = 22/57 (38%), Positives = 29/57 (50%), Gaps = 5/57 (8%)
Frame = -3
Query: 2 PLSSTSDCELLCGEDSSEVL---TGDLPECSS--DLDSSSSSQLPSSSLFAEEEEES 53
P ST+ +SSE L G+ P SS DL+S S S+ SSS+ + EE S
Sbjct: 357 PSESTTSSSSS*ESESSEAL**VVGEAPSSSSNSDLESESESEESSSSMLSAEESSS 187
>TC82655 similar to PIR|T49930|T49930 hypothetical protein F17I14.130 -
Arabidopsis thaliana, partial (45%)
Length = 719
Score = 30.4 bits (67), Expect = 1.0
Identities = 21/76 (27%), Positives = 39/76 (50%)
Frame = -1
Query: 131 LLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILDWRLRSITPLS 190
L+ + + + K +EP+VPS+L+F+ K +P LL+L +L + S +S
Sbjct: 410 LVDLLAVLVTLKDKEPIVPSILEFE----KSFDKPSFHKTPPLLLLLLLAFEDSSFMEIS 243
Query: 191 FLSFFACKLDSTGTFT 206
+S + + +TG T
Sbjct: 242 AISETSSFVSTTGVLT 195
>BQ137360
Length = 978
Score = 30.4 bits (67), Expect = 1.0
Identities = 9/22 (40%), Positives = 14/22 (62%)
Frame = +1
Query: 251 FVNPEHAESWCEGLSKEKIIGC 272
F NP + WC+ LS+ ++GC
Sbjct: 268 FANPHVSPQWCDSLSRTSLVGC 333
>TC93240 weakly similar to GP|14335170|gb|AAK59865.1 AT5g51070/K3K7_27
{Arabidopsis thaliana}, partial (8%)
Length = 700
Score = 29.6 bits (65), Expect = 1.8
Identities = 19/38 (50%), Positives = 22/38 (57%)
Frame = +1
Query: 293 PQLRVTARTRRWSTVSSSSSSPSSSSSPSFSLSYKKRK 330
P L + R S+ SSSSSS SSSSS SL+ K K
Sbjct: 166 PSLSFLSSQRNRSSSSSSSSSSSSSSSSFTSLTSTKTK 279
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.131 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,275,696
Number of Sequences: 36976
Number of extensions: 200396
Number of successful extensions: 3566
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 2117
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2908
length of query: 345
length of database: 9,014,727
effective HSP length: 97
effective length of query: 248
effective length of database: 5,428,055
effective search space: 1346157640
effective search space used: 1346157640
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146723.15


