

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146719.4 - phase: 0 /pseudo
(471 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
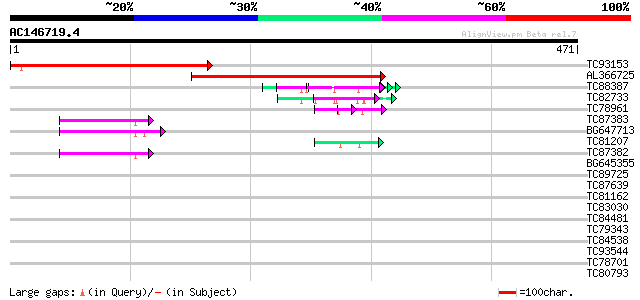
Score E
Sequences producing significant alignments: (bits) Value
TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Ci... 284 6e-77
AL366725 273 1e-73
TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finge... 53 2e-07
TC82733 similar to GP|10177404|dbj|BAB10535. gene_id:K24M7.12~pi... 49 4e-06
TC78961 similar to GP|18252179|gb|AAL61922.1 unknown protein {Ar... 49 5e-06
TC87383 similar to GP|19168656|emb|CAD26175. DNA-DIRECTED RNA PO... 47 1e-05
BG647713 homologue to GP|15042313|gb| 232R {Chilo iridescent vir... 47 2e-05
TC81207 similar to GP|21322752|dbj|BAB78536. cold shock protein-... 46 3e-05
TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulch... 45 6e-05
BG645355 similar to PIR|G96590|G965 hypothetical protein T24C10.... 34 0.11
TC89725 similar to PIR|T05494|T05494 glycine-rich protein T19K4.... 32 0.40
TC87639 GP|9663153|emb|CAC01132.1 transport-secretion protein 2.... 32 0.53
TC81162 weakly similar to PIR|T47673|T47673 hypothetical protein... 31 0.90
TC83030 weakly similar to GP|18855061|gb|AAL79753.1 putative RNA... 31 0.90
TC84481 similar to GP|11875626|gb|AAG40731.1 XRN4 {Arabidopsis t... 30 2.0
TC79343 weakly similar to PIR|E86411|E86411 protein F1K23.18 [im... 30 2.0
TC84538 weakly similar to GP|22296418|dbj|BAC10186. similar to p... 30 2.6
TC93544 weakly similar to SP|O82256|COLA_ARATH Zinc finger prote... 29 3.4
TC78701 similar to GP|21554835|gb|AAM63703.1 unknown {Arabidopsi... 29 3.4
TC80793 similar to SP|Q9LUA9|COLE_ARATH Zinc finger protein cons... 29 3.4
>TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Cicer
arietinum}, partial (8%)
Length = 516
Score = 284 bits (726), Expect = 6e-77
Identities = 136/171 (79%), Positives = 148/171 (86%), Gaps = 3/171 (1%)
Frame = +2
Query: 1 MAGRNDAA---ALKAIAQAVGQQPNAGAGNDGVRMLETFLRNHPPTFKGRYDPDGAQKWL 57
M G +DAA AL+A+AQAV Q P G+DG RMLETFLRNHPPTFKGRY PDGA KWL
Sbjct: 2 MTGSSDAALVAALEAVAQAVQQLPKVDTGSDGTRMLETFLRNHPPTFKGRYAPDGA*KWL 181
Query: 58 KEVERIFRVMQCSEVQKVRFGTHMLAEEADDWWVSLLPILEQDGAAVTWVVFRREFLNMY 117
KE+ERIFRVMQC E QKV+FGTHMLAEEADDWW+SLLP+LEQD A VTW +FR+EFL Y
Sbjct: 182 KEIERIFRVMQCFETQKVQFGTHMLAEEADDWWISLLPVLEQDDAVVTWAMFRKEFLGRY 361
Query: 118 FPEDVRGKKEIEFLEMKQGDMSVTEYAAKFVELAKFYPHYTTETAEFSKCI 168
FPEDVRGKKEIEFLE+KQGDMSVTEYAAKFVELA FYPHY+ ETAEFSKCI
Sbjct: 362 FPEDVRGKKEIEFLELKQGDMSVTEYAAKFVELATFYPHYSAETAEFSKCI 514
>AL366725
Length = 485
Score = 273 bits (697), Expect = 1e-73
Identities = 120/161 (74%), Positives = 142/161 (87%)
Frame = +2
Query: 152 KFYPHYTTETAEFSKCIKFENGLRADIKRAIGYEKIRIFSELVSSCRIYEEDTKAHYKVM 211
KFYPHY ETAEFSKCIKFENGLR DIKRAIGY+++R+F +LV++CRIYEEDTKAH KV+
Sbjct: 2 KFYPHYAAETAEFSKCIKFENGLRPDIKRAIGYQQLRVFPDLVNTCRIYEEDTKAHDKVV 181
Query: 212 SERRGKGQ*SRPKPYSAPTDKGKQRLNDERRPKRRDAPTEIICFKCGEKGHKSNVCDRDE 271
+ER+ KGQ*SRPKPYSAP DKGKQR+ D+RRPK++DAP EI+CF GEKGHKSNVC ++
Sbjct: 182 NERKTKGQ*SRPKPYSAPADKGKQRMVDDRRPKKKDAPAEIVCFNYGEKGHKSNVCPKEI 361
Query: 272 KKCFRCGQKGHTLADCKRGDVVCYNCNEEGHISSQCTQPKK 312
KKC RC +KGH +ADCKR D+VC+NCNEEGHI SQC QPK+
Sbjct: 362 KKCVRCDKKGHIVADCKRNDIVCFNCNEEGHIGSQCKQPKR 484
>TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finger protein
{Oryza sativa (japonica cultivar-group)}, partial (96%)
Length = 1286
Score = 53.1 bits (126), Expect = 2e-07
Identities = 27/96 (28%), Positives = 46/96 (47%), Gaps = 5/96 (5%)
Frame = +1
Query: 222 RPKPYSAPTDKGKQRLNDERRPKR-----RDAPTEIICFKCGEKGHKSNVCDRDEKKCFR 276
R PY + +G + N + KR R+ P +C C GH ++ C + C+
Sbjct: 370 REAPYRRDSRRGFSQDNLCKNCKRPGHYVRECPNVAVCHNCSLPGHIASECST-KSLCWN 546
Query: 277 CGQKGHTLADCKRGDVVCYNCNEEGHISSQCTQPKK 312
C + GH + C + +C+ C + GH + +CT P+K
Sbjct: 547 CKEPGHMASSCPN-EGICHTCGKAGHRARECTVPQK 651
Score = 52.8 bits (125), Expect = 3e-07
Identities = 25/71 (35%), Positives = 40/71 (56%), Gaps = 6/71 (8%)
Frame = +1
Query: 247 DAPTEIICFKCGEKGHKSNVCDRDEKKCFRCGQKGHTLADCK-----RGDV-VCYNCNEE 300
+ T+ +C+ C E GH ++ C +E C CG+ GH +C GD+ +C NC ++
Sbjct: 517 ECSTKSLCWNCKEPGHMASSCP-NEGICHTCGKAGHRARECTVPQKPPGDLRLCNNCYKQ 693
Query: 301 GHISSQCTQPK 311
GHI+ +CT K
Sbjct: 694 GHIAVECTNEK 726
Score = 49.3 bits (116), Expect = 3e-06
Identities = 32/117 (27%), Positives = 45/117 (38%), Gaps = 9/117 (7%)
Frame = +1
Query: 211 MSERRGKGQ*SRPKPYSAPTDKGKQRLNDER---------RPKRRDAPTEIICFKCGEKG 261
+S R K + P S +R+ ER R RR + +C C G
Sbjct: 268 VSRSRSKSRSRSPMDRSRSRSPVDRRIRSERFSHREAPYRRDSRRGFSQDNLCKNCKRPG 447
Query: 262 HKSNVCDRDEKKCFRCGQKGHTLADCKRGDVVCYNCNEEGHISSQCTQPKKVRTGGK 318
H C + C C GH ++C + C+NC E GH++S C T GK
Sbjct: 448 HYVRECP-NVAVCHNCSLPGHIASECSTKSL-CWNCKEPGHMASSCPNEGICHTCGK 612
Score = 48.1 bits (113), Expect = 7e-06
Identities = 28/101 (27%), Positives = 39/101 (37%), Gaps = 25/101 (24%)
Frame = +1
Query: 249 PTEIICFKCGEKGHKSNVCD-------------------------RDEKKCFRCGQKGHT 283
P E IC CG+ GH++ C +EK C C + GH
Sbjct: 580 PNEGICHTCGKAGHRARECTVPQKPPGDLRLCNNCYKQGHIAVECTNEKACNNCRKTGHL 759
Query: 284 LADCKRGDVVCYNCNEEGHISSQCTQPKKVRTGGKVFALTG 324
DC D +C CN GH++ QC + + G +L G
Sbjct: 760 ARDCPN-DPICNLCNISGHVARQCPKSNVIGDRGGGGSLRG 879
Score = 40.0 bits (92), Expect = 0.002
Identities = 23/85 (27%), Positives = 31/85 (36%), Gaps = 23/85 (27%)
Frame = +1
Query: 246 RDAPTEIICFKCGEKGHKSNVCDRD----------------------EKKCFRCGQKGHT 283
RD P + IC C GH + C + + C C Q GH
Sbjct: 763 RDCPNDPICNLCNISGHVARQCPKSNVIGDRGGGGSLRGGYRDGGFRDVVCRSCQQFGHM 942
Query: 284 LADCKRGDV-VCYNCNEEGHISSQC 307
DC G + +C NC GH + +C
Sbjct: 943 SRDCMGGPLMICQNCGGRGHQAYEC 1017
Score = 30.8 bits (68), Expect = 1.2
Identities = 14/44 (31%), Positives = 20/44 (44%), Gaps = 1/44 (2%)
Frame = +1
Query: 251 EIICFKCGEKGHKSNVCDRDEKK-CFRCGQKGHTLADCKRGDVV 293
+++C C + GH S C C CG +GH +C G V
Sbjct: 904 DVVCRSCQQFGHMSRDCMGGPLMICQNCGGRGHQAYECPSGRFV 1035
>TC82733 similar to GP|10177404|dbj|BAB10535.
gene_id:K24M7.12~pir||S42136~similar to unknown protein
{Arabidopsis thaliana}, partial (57%)
Length = 710
Score = 48.9 bits (115), Expect = 4e-06
Identities = 23/67 (34%), Positives = 35/67 (51%), Gaps = 12/67 (17%)
Frame = +3
Query: 253 ICFKCGEKGHKSNVCD-----RDEKKCFRCGQKGHTLADC----KRGDVV---CYNCNEE 300
IC +C +GH++ C D K + CG GH+LA+C + G + C+ C E+
Sbjct: 354 ICLRCRRRGHRAQNCPDGGSKEDFKY*YNCGDNGHSLANCPHPLQEGGTMFAQCFVCKEQ 533
Query: 301 GHISSQC 307
GH+S C
Sbjct: 534 GHLSKNC 554
Score = 44.3 bits (103), Expect = 1e-04
Identities = 31/121 (25%), Positives = 49/121 (39%), Gaps = 22/121 (18%)
Frame = +3
Query: 223 PKPYSAPTDKGKQRLNDER-----RPKRRDAPTEI-------ICFKCGEKGHKSNVCDRD 270
P P + P+ K K + ++ +P+ P + CF C H + C +
Sbjct: 153 PTPPNDPSKKKKNKFKRKKPDSNSKPRTGKRPLRVPGMKPGDSCFICKGLDHIAKFCTQK 332
Query: 271 -----EKKCFRCGQKGHTLADCKRGDV-----VCYNCNEEGHISSQCTQPKKVRTGGKVF 320
K C RC ++GH +C G YNC + GH + C P ++ GG +F
Sbjct: 333 AEWEKNKICLRCRRRGHRAQNCPDGGSKEDFKY*YNCGDNGHSLANCPHP--LQEGGTMF 506
Query: 321 A 321
A
Sbjct: 507 A 509
>TC78961 similar to GP|18252179|gb|AAL61922.1 unknown protein {Arabidopsis
thaliana}, partial (71%)
Length = 974
Score = 48.5 bits (114), Expect = 5e-06
Identities = 22/44 (50%), Positives = 24/44 (54%), Gaps = 3/44 (6%)
Frame = +2
Query: 273 KCFRCGQKGHTLADCKRGD--VVCYNCNEEGHISSQC-TQPKKV 313
+CF CG GH DCK GD CY C E GHI C PKK+
Sbjct: 356 RCFNCGIDGHWARDCKAGDWKNKCYRCGERGHIEKNCKNSPKKL 487
Score = 41.2 bits (95), Expect = 9e-04
Identities = 16/37 (43%), Positives = 22/37 (59%), Gaps = 2/37 (5%)
Frame = +2
Query: 254 CFKCGEKGHKSNVCDRDE--KKCFRCGQKGHTLADCK 288
CF CG GH + C + KC+RCG++GH +CK
Sbjct: 359 CFNCGIDGHWARDCKAGDWKNKCYRCGERGHIEKNCK 469
Score = 28.9 bits (63), Expect = 4.4
Identities = 10/23 (43%), Positives = 13/23 (56%)
Frame = +2
Query: 254 CFKCGEKGHKSNVCDRDEKKCFR 276
C++CGE+GH C KK R
Sbjct: 425 CYRCGERGHIEKNCKNSPKKLSR 493
>TC87383 similar to GP|19168656|emb|CAD26175. DNA-DIRECTED RNA POLYMERASE II
{Encephalitozoon cuniculi}, partial (0%)
Length = 1247
Score = 47.4 bits (111), Expect = 1e-05
Identities = 24/80 (30%), Positives = 40/80 (50%), Gaps = 2/80 (2%)
Frame = -2
Query: 42 PTFKGRYDPDGAQKWLKEVERIFRVMQCSEVQKVRFGTHMLAEEADDWWVSLLPILEQDG 101
P F+G PD WL+ +ER+F+ + E QKV+ L + A WW ++ +++G
Sbjct: 724 PDFEGNLQPDDLLDWLQIMERLFKYKEVLEEQKVKIVAAKLKKLASIWWENVKRRRKREG 545
Query: 102 AA--VTWVVFRREFLNMYFP 119
+ TW R++ Y P
Sbjct: 544 KSKIKTWEKMRQKLTRKYLP 485
>BG647713 homologue to GP|15042313|gb| 232R {Chilo iridescent virus}, partial
(1%)
Length = 726
Score = 46.6 bits (109), Expect = 2e-05
Identities = 29/95 (30%), Positives = 48/95 (50%), Gaps = 7/95 (7%)
Frame = +2
Query: 42 PTFKGRYDPDGAQKWLKEVERIFRVMQCSEVQKVRFGTHMLAEEADDWWVSLLPILEQDG 101
P F G+ PD WL+ +ER+F+ + +E QKV+ L + A WW +L +G
Sbjct: 347 PDF*GKLQPDEFVDWLQTIERVFKYKEVAEEQKVKIVAAKLKKHASIWWKNLKRKRNCEG 526
Query: 102 AA--VTWVVFR----REFLN-MYFPEDVRGKKEIE 129
+ TW R R++L+ Y+ ++ KK I+
Sbjct: 527 KSKIKTWDKMRQKLTRKYLHPHYYQDNFTQKKNIQ 631
>TC81207 similar to GP|21322752|dbj|BAB78536. cold shock protein-1 {Triticum
aestivum}, partial (39%)
Length = 630
Score = 46.2 bits (108), Expect = 3e-05
Identities = 24/88 (27%), Positives = 31/88 (34%), Gaps = 31/88 (35%)
Frame = +3
Query: 254 CFKCGEKGHKSNVCDRDEKK-----------------CFRCGQKGHTLADCKR------- 289
C+ CG+ GH + CDR ++ C+ CG H DC R
Sbjct: 351 CYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNN 530
Query: 290 -------GDVVCYNCNEEGHISSQCTQP 310
G CY C GHI+ C P
Sbjct: 531 GGGGYGGGGTSCYRCGGVGHIARDCATP 614
Score = 27.7 bits (60), Expect = 9.9
Identities = 13/44 (29%), Positives = 21/44 (47%), Gaps = 4/44 (9%)
Frame = +3
Query: 278 GQKGHTLADCKRGDVVCYNCNEEGHISSQCTQ----PKKVRTGG 317
G +G + + G CY C + GHI+ C + + R+GG
Sbjct: 303 GGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGG 434
>TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulchellus},
partial (7%)
Length = 2304
Score = 45.1 bits (105), Expect = 6e-05
Identities = 24/80 (30%), Positives = 38/80 (47%), Gaps = 2/80 (2%)
Frame = +2
Query: 42 PTFKGRYDPDGAQKWLKEVERIFRVMQCSEVQKVRFGTHMLAEEADDWWVSLLPILEQDG 101
P F+G D WL+ +ER+F + E QKV+ L + A WW +L +++G
Sbjct: 629 PDFEGNLQLDDFLDWLQTIERVFEYKEVPEEQKVKIVAAKLKKHALIWWENLKRRRKREG 808
Query: 102 AA--VTWVVFRREFLNMYFP 119
+ TW R++ Y P
Sbjct: 809 KSKIKTWDKMRQKLTRKYLP 868
>BG645355 similar to PIR|G96590|G965 hypothetical protein T24C10.5 [imported]
- Arabidopsis thaliana, partial (5%)
Length = 627
Score = 34.3 bits (77), Expect = 0.11
Identities = 14/48 (29%), Positives = 21/48 (43%), Gaps = 13/48 (27%)
Frame = -1
Query: 273 KCFRCGQKGHTLADCKRGDVV-------------CYNCNEEGHISSQC 307
KC++C Q GH ++C CY CN+ GH ++ C
Sbjct: 504 KCYKCQQPGHWASNCPSMSAANRVSGGSGGASGNCYKCNQPGHWANNC 361
>TC89725 similar to PIR|T05494|T05494 glycine-rich protein T19K4.150 -
Arabidopsis thaliana, partial (17%)
Length = 378
Score = 32.3 bits (72), Expect = 0.40
Identities = 14/54 (25%), Positives = 19/54 (34%), Gaps = 20/54 (37%)
Frame = +1
Query: 274 CFRCGQKGHTLADCKRGDV--------------------VCYNCNEEGHISSQC 307
C+ CG+ GH +C G CY+C E GH + C
Sbjct: 16 CYNCGESGHMARECTSGGGGGGGRYGGGGGGGGGGGGGGSCYSCGESGHFARDC 177
Score = 32.3 bits (72), Expect = 0.40
Identities = 15/57 (26%), Positives = 19/57 (33%), Gaps = 20/57 (35%)
Frame = +1
Query: 254 CFKCGEKGHKSNVCDRDEK--------------------KCFRCGQKGHTLADCKRG 290
C+ CGE GH + C C+ CG+ GH DC G
Sbjct: 16 CYNCGESGHMARECTSGGGGGGGRYGGGGGGGGGGGGGGSCYSCGESGHFARDCPTG 186
Score = 30.4 bits (67), Expect = 1.5
Identities = 9/15 (60%), Positives = 12/15 (80%)
Frame = +1
Query: 294 CYNCNEEGHISSQCT 308
CYNC E GH++ +CT
Sbjct: 16 CYNCGESGHMARECT 60
>TC87639 GP|9663153|emb|CAC01132.1 transport-secretion protein 2.2 (TTS-2.2)
{Homo sapiens}, partial (2%)
Length = 1522
Score = 32.0 bits (71), Expect = 0.53
Identities = 14/29 (48%), Positives = 18/29 (61%), Gaps = 3/29 (10%)
Frame = -1
Query: 294 CYNCNEEGHISSQCTQ---PKKVRTGGKV 319
CY+C+E GHI+ CT KK + G KV
Sbjct: 316 CYSCHERGHIARNCTNTSAAKKKKKGPKV 230
>TC81162 weakly similar to PIR|T47673|T47673 hypothetical protein T26I12.220
- Arabidopsis thaliana, partial (9%)
Length = 756
Score = 31.2 bits (69), Expect = 0.90
Identities = 11/29 (37%), Positives = 19/29 (64%)
Frame = +2
Query: 283 TLADCKRGDVVCYNCNEEGHISSQCTQPK 311
++A KR + +CY C ++GH S+C P+
Sbjct: 389 SVASGKRKNRMCYGCRQKGHNLSECPNPQ 475
>TC83030 weakly similar to GP|18855061|gb|AAL79753.1 putative RNA helicase
{Oryza sativa}, partial (7%)
Length = 624
Score = 31.2 bits (69), Expect = 0.90
Identities = 23/64 (35%), Positives = 28/64 (42%), Gaps = 8/64 (12%)
Frame = +1
Query: 212 SERRG----KGQ*SRPKPYSAPTDK----GKQRLNDERRPKRRDAPTEIICFKCGEKGHK 263
S RRG K S KP + D G+Q P R A T CF CGE GH+
Sbjct: 25 SSRRGGRSYKSGNSWSKPERSSRDDWLIGGRQSSRSSSSPNRSFAGT---CFTCGESGHR 195
Query: 264 SNVC 267
++ C
Sbjct: 196 ASDC 207
Score = 30.8 bits (68), Expect = 1.2
Identities = 12/20 (60%), Positives = 14/20 (70%), Gaps = 2/20 (10%)
Frame = +1
Query: 274 CFRCGQKGHTLADC--KRGD 291
CF CG+ GH +DC KRGD
Sbjct: 166 CFTCGESGHRASDCPNKRGD 225
>TC84481 similar to GP|11875626|gb|AAG40731.1 XRN4 {Arabidopsis thaliana},
partial (23%)
Length = 727
Score = 30.0 bits (66), Expect = 2.0
Identities = 9/18 (50%), Positives = 14/18 (77%)
Frame = +2
Query: 271 EKKCFRCGQKGHTLADCK 288
++KC+ CGQ GH A+C+
Sbjct: 200 QEKCYMCGQVGHLAAECR 253
>TC79343 weakly similar to PIR|E86411|E86411 protein F1K23.18 [imported] -
Arabidopsis thaliana, partial (52%)
Length = 1420
Score = 30.0 bits (66), Expect = 2.0
Identities = 25/96 (26%), Positives = 42/96 (43%), Gaps = 7/96 (7%)
Frame = +3
Query: 10 LKAIAQAVGQQPNAGA------GNDGVRMLETFLRNHPPTFKGRYDPDGAQKWLKEVERI 63
+ AI A+ + + GA GN + +L + T K +YD G KWL E
Sbjct: 708 ISAITSAINELIDLGARTLMIPGNFPLGCNVIYLTKYETTDKSQYDSAGCLKWLNEFAEF 887
Query: 64 FRVMQCSEVQKV-RFGTHMLAEEADDWWVSLLPILE 98
+ E+ ++ R H A D++ +LLP+ +
Sbjct: 888 YNQELQYELHRLRRIHPHATIIYA-DYYNALLPLYQ 992
>TC84538 weakly similar to GP|22296418|dbj|BAC10186. similar to pre-mRNA
splicing factor ATP-dependent RNA helicase, partial
(11%)
Length = 973
Score = 29.6 bits (65), Expect = 2.6
Identities = 16/41 (39%), Positives = 19/41 (46%), Gaps = 5/41 (12%)
Frame = +2
Query: 272 KKCFRCGQ-----KGHTLADCKRGDVVCYNCNEEGHISSQC 307
K CF CGQ G +CK G VC+ C E S +C
Sbjct: 764 KSCFACGQIIEKIDGCNHVECKCGKHVCWVCLEIFTSSDEC 886
>TC93544 weakly similar to SP|O82256|COLA_ARATH Zinc finger protein
constans-like 10. [Mouse-ear cress] {Arabidopsis
thaliana}, partial (45%)
Length = 731
Score = 29.3 bits (64), Expect = 3.4
Identities = 14/53 (26%), Positives = 20/53 (37%), Gaps = 3/53 (5%)
Frame = +2
Query: 253 ICFKCGEKGHKSNVCDRDEKK---CFRCGQKGHTLADCKRGDVVCYNCNEEGH 302
+CF C + H +N + C C T+ V C NC+ E H
Sbjct: 122 LCFSCDREVHSTNQLFSKHTRSLICDSCDDSPATILCSTESSVFCQNCDWENH 280
>TC78701 similar to GP|21554835|gb|AAM63703.1 unknown {Arabidopsis
thaliana}, partial (44%)
Length = 1480
Score = 29.3 bits (64), Expect = 3.4
Identities = 18/87 (20%), Positives = 35/87 (39%)
Frame = -1
Query: 296 NCNEEGHISSQCTQPKKVRTGGKVFALTGTQTVNEDRLIRGTCFFNSTHLIAIIDTGATH 355
+CN I + T PK + + KVF T T +++ + + II G +
Sbjct: 358 SCNGTERILTDSTSPKTLNSFNKVFFFTLTGKFPTKIVLKSLSSSSKPSFVTIISAGLSR 179
Query: 356 CFIVVDCAYKLGLIVSDMNGEMVVETP 382
F ++ + +S N +++ P
Sbjct: 178 HFSLIPNFLHTFIFISSFNNPILLLLP 98
>TC80793 similar to SP|Q9LUA9|COLE_ARATH Zinc finger protein constans-like
14. [Mouse-ear cress] {Arabidopsis thaliana}, partial
(27%)
Length = 937
Score = 29.3 bits (64), Expect = 3.4
Identities = 18/64 (28%), Positives = 27/64 (42%), Gaps = 4/64 (6%)
Frame = +1
Query: 253 ICFKCGEKGHKSNVCDRDEKK---CFRCGQKGHTLADCKRGDV-VCYNCNEEGHISSQCT 308
+C C H +N R + C RC + L C V +C NC+ GH +S +
Sbjct: 307 LCLSCDRNVHSANTLARRHSRTLLCERCSSQP-ALVRCSEEKVSLCQNCDWLGHGNSTSS 483
Query: 309 QPKK 312
K+
Sbjct: 484 NHKR 495
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.137 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,390,435
Number of Sequences: 36976
Number of extensions: 257098
Number of successful extensions: 1264
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 1161
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1231
length of query: 471
length of database: 9,014,727
effective HSP length: 100
effective length of query: 371
effective length of database: 5,317,127
effective search space: 1972654117
effective search space used: 1972654117
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146719.4


