

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146711.3 - phase: 0
(61 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
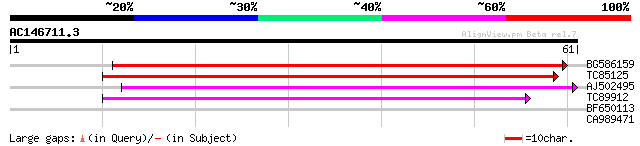
BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2... 52 5e-08
TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-relate... 47 1e-06
AJ502495 weakly similar to GP|18071369|g putative gag-pol polypr... 45 4e-06
TC89912 weakly similar to PIR|B84512|B84512 probable retroelemen... 43 3e-05
BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vu... 36 0.003
CA989471 similar to GP|11994367|dbj gb|AAF17567.1~gene_id:MDC16.... 25 6.0
>BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (11%)
Length = 732
Score = 52.0 bits (123), Expect = 5e-08
Identities = 25/49 (51%), Positives = 32/49 (65%)
Frame = +1
Query: 12 DADYAGDKIERKSTSGSCQFLEQALTGWSCRKQNTIALSTTEAEYESAA 60
D+DYAGD +RKSTSG L WS +KQ + LSTT+AE+ +AA
Sbjct: 376 DSDYAGDLDDRKSTSGYVFMLSSGAVSWSSKKQPVVTLSTTKAEFIAAA 522
>TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
(EC 3.4.23.-);, partial (7%)
Length = 705
Score = 47.0 bits (110), Expect = 1e-06
Identities = 23/49 (46%), Positives = 31/49 (62%)
Frame = +1
Query: 11 LDADYAGDKIERKSTSGSCQFLEQALTGWSCRKQNTIALSTTEAEYESA 59
+D+D+AGD +RKST+G L W + Q +ALSTTEAEY +A
Sbjct: 85 VDSDFAGDHDKRKSTTGYVFTLAGGAVSWLSKLQTVVALSTTEAEYMAA 231
>AJ502495 weakly similar to GP|18071369|g putative gag-pol polyprotein
{Oryza sativa}, partial (9%)
Length = 542
Score = 45.4 bits (106), Expect = 4e-06
Identities = 23/49 (46%), Positives = 29/49 (58%)
Frame = +2
Query: 13 ADYAGDKIERKSTSGSCQFLEQALTGWSCRKQNTIALSTTEAEYESAAS 61
+D+AGD RKSTSG L WS +KQ +A ST EAEY ++ S
Sbjct: 2 SDWAGDTETRKSTSGYAFHLGTGAISWSSKKQPVVAFSTAEAEYIASTS 148
>TC89912 weakly similar to PIR|B84512|B84512 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(10%)
Length = 814
Score = 42.7 bits (99), Expect = 3e-05
Identities = 21/46 (45%), Positives = 27/46 (58%)
Frame = +1
Query: 11 LDADYAGDKIERKSTSGSCQFLEQALTGWSCRKQNTIALSTTEAEY 56
+DADYAG+ RKS SG L W +Q+ + LSTT+AEY
Sbjct: 100 VDADYAGNVDTRKSLSGFVFTLYGTTISWKANQQSVVTLSTTQAEY 237
>BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vulgaris},
partial (13%)
Length = 494
Score = 35.8 bits (81), Expect = 0.003
Identities = 20/45 (44%), Positives = 25/45 (55%)
Frame = +1
Query: 12 DADYAGDKIERKSTSGSCQFLEQALTGWSCRKQNTIALSTTEAEY 56
D+D+ GD R+STSG A W +KQ ALS+ EAEY
Sbjct: 295 DSDWCGD---RRSTSGYVFKFNDAAISWCTKKQPITALSSYEAEY 420
>CA989471 similar to GP|11994367|dbj gb|AAF17567.1~gene_id:MDC16.8~similar to
unknown protein {Arabidopsis thaliana}, partial (7%)
Length = 652
Score = 25.0 bits (53), Expect = 6.0
Identities = 13/42 (30%), Positives = 20/42 (46%)
Frame = -3
Query: 8 CSLLDADYAGDKIERKSTSGSCQFLEQALTGWSCRKQNTIAL 49
CSL+ D++ + + C L LT C KQN++ L
Sbjct: 527 CSLVSTHQRNDEVVSELSHFLCPCLLFLLT*CQCLKQNSVVL 402
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.332 0.136 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,746,436
Number of Sequences: 36976
Number of extensions: 16436
Number of successful extensions: 129
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 129
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 129
length of query: 61
length of database: 9,014,727
effective HSP length: 37
effective length of query: 24
effective length of database: 7,646,615
effective search space: 183518760
effective search space used: 183518760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 51 (24.3 bits)
Medicago: description of AC146711.3


