

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146709.12 + phase: 0 /pseudo
(609 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
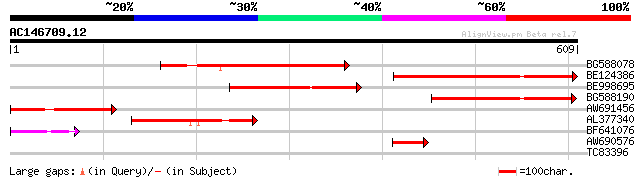
Score E
Sequences producing significant alignments: (bits) Value
BG588078 similar to GP|4314359|gb| unknown protein {Arabidopsis ... 317 8e-87
BE124386 weakly similar to GP|16323222|gb AT4g24430/T22A6_260 {A... 223 2e-58
BE998695 similar to GP|16323222|gb AT4g24430/T22A6_260 {Arabidop... 158 5e-39
BG588190 similar to GP|2160180|gb| F21M12.30 gene product {Arabi... 156 2e-38
AW691456 123 2e-28
AL377340 similar to GP|2160178|gb| F21M12.27 gene product {Arabi... 115 4e-26
BF641076 similar to GP|2160180|gb|A F21M12.30 gene product {Arab... 60 3e-09
AW690576 similar to GP|16323222|gb| AT4g24430/T22A6_260 {Arabido... 58 9e-09
TC83396 similar to GP|22858664|gb|AAN05792.1 unknown {Gossypium ... 29 6.0
>BG588078 similar to GP|4314359|gb| unknown protein {Arabidopsis thaliana},
partial (20%)
Length = 623
Score = 317 bits (812), Expect = 8e-87
Identities = 161/216 (74%), Positives = 171/216 (78%), Gaps = 13/216 (6%)
Frame = +2
Query: 163 VLLTNPINPQFRGEVDDKYQYSCENKDTQFMDGLVLILIHQWVSG**HRVMSFAMVDPLS 222
VLLTNPINPQFR EVDDKYQYS EN+D +H W+ + F M+ P +
Sbjct: 2 VLLTNPINPQFRLEVDDKYQYSWENRDNT---------VHGWIGFDSDPPVGFWMITPSN 154
Query: 223 KIL------------LLMLDQSLF-RTHYAGKEVTMAFQEGETYKKVFGPVFVYLNTASS 269
+ + + S+F THYAGKEVTMAFQEGETYKKVFGPVFVYLNTASS
Sbjct: 155 EFRNGGPIKQDLTSHVGPIALSMFVSTHYAGKEVTMAFQEGETYKKVFGPVFVYLNTASS 334
Query: 270 ENDNATLWSDAVQQLSKEVQSWPYDFPQSQDYFPPNQRGAVFGRLLVQDWYFEGGRYQYA 329
ENDNATLWSDAVQQLSKEVQSWPYDFPQSQDYFPPNQRGAVFGRLLVQDWYFEGGRYQYA
Sbjct: 335 ENDNATLWSDAVQQLSKEVQSWPYDFPQSQDYFPPNQRGAVFGRLLVQDWYFEGGRYQYA 514
Query: 330 NNAYVGLALPGDAGSWQRESKGYQFWTQTDAKGYFK 365
NNAYVGLALPGDAGSWQRESKGYQFWTQTDAKGYFK
Sbjct: 515 NNAYVGLALPGDAGSWQRESKGYQFWTQTDAKGYFK 622
>BE124386 weakly similar to GP|16323222|gb AT4g24430/T22A6_260 {Arabidopsis
thaliana}, partial (37%)
Length = 634
Score = 223 bits (567), Expect = 2e-58
Identities = 109/201 (54%), Positives = 143/201 (70%), Gaps = 4/201 (1%)
Frame = +2
Query: 413 NGPTIWEIGIPDRLTSEFHVPEPYPTLMNKLYTEGRDK--QYGLWERYTEMYPTDDLIYT 470
+GPT+WEIGIPDR +EF+VP+P P +N+LY D+ QYGLWERY ++YP +DL+YT
Sbjct: 5 HGPTLWEIGIPDRSAAEFYVPDPNPLYVNRLYVNHSDRFRQYGLWERYADLYPNEDLVYT 184
Query: 471 LGVNK-NKDWFYAHVTRNTENNTYEPTTWQIIFEHQHDLKSGNYTLQLALASAADAYLQV 529
+G++ KDWF+A VTR +NNTY+ TTW+I F G Y L+LALAS A LQ+
Sbjct: 185 VGISDYKKDWFFAQVTRKKDNNTYQGTTWRINFNLDSVNTKGTYKLRLALASVHYAELQI 364
Query: 530 RFNDR-SVYPPHFATGHIGRIRGDNCIQRHGIHGLYRLFSIDVPSNLLLKGKNIIYLTQT 588
R N+ + PP F+TG IG+ +N I RHGIHGLY L++IDV +LL+KG N I+LTQ
Sbjct: 365 RINNLDATSPPLFSTGVIGK---ENTIARHGIHGLYWLYNIDVQGSLLVKGNNTIFLTQA 535
Query: 589 NADTPFQGVMYDYIRLEQPPS 609
D PF G++YDYIRLE PP+
Sbjct: 536 TKDGPFVGILYDYIRLEGPPT 598
>BE998695 similar to GP|16323222|gb AT4g24430/T22A6_260 {Arabidopsis
thaliana}, partial (31%)
Length = 583
Score = 158 bits (400), Expect = 5e-39
Identities = 77/143 (53%), Positives = 99/143 (68%), Gaps = 2/143 (1%)
Frame = +2
Query: 237 HYAGKEVTMAFQEGETYKKVFGPVFVYLNTASSENDN--ATLWSDAVQQLSKEVQSWPYD 294
HYAG+++ + Q E +KKVFGP FVYLN +++ A LW DA Q++KEVQSWPYD
Sbjct: 155 HYAGEDIVLKLQPNEPWKKVFGPTFVYLNNLLDHDEDPLAQLWEDAKFQMNKEVQSWPYD 334
Query: 295 FPQSQDYFPPNQRGAVFGRLLVQDWYFEGGRYQYANNAYVGLALPGDAGSWQRESKGYQF 354
FP S D+ +QRG+V G LLV+D + A AYVGLA PGDAGSWQRE KGYQF
Sbjct: 335 FPASDDFQKASQRGSVCGTLLVRDRCVSD-KDIIAKGAYVGLAPPGDAGSWQRECKGYQF 511
Query: 355 WTQTDAKGYFKITNVVPGHYNLF 377
W++++ +GYF I N+ G YNL+
Sbjct: 512 WSKSNEEGYFSINNIRSGDYNLY 580
>BG588190 similar to GP|2160180|gb| F21M12.30 gene product {Arabidopsis
thaliana}, partial (15%)
Length = 547
Score = 156 bits (395), Expect = 2e-38
Identities = 81/156 (51%), Positives = 104/156 (65%), Gaps = 1/156 (0%)
Frame = +2
Query: 454 LWERYTEMYPTDDLIYTLGVNK-NKDWFYAHVTRNTENNTYEPTTWQIIFEHQHDLKSGN 512
L + YTE+YP +L+YT+GV+ KDWFYA V R +NT E TTWQI FE ++
Sbjct: 2 LLDIYTELYPDANLVYTIGVSDYRKDWFYAQVPRKKADNTLEGTTWQIKFELSGVIQGAT 181
Query: 513 YTLQLALASAADAYLQVRFNDRSVYPPHFATGHIGRIRGDNCIQRHGIHGLYRLFSIDVP 572
Y L++A+ASA A LQ+R ND + P F TG IGR +N I R GIHGLY L+ +++P
Sbjct: 182 YKLRVAIASATLAELQIRVNDPNARRPVFTTGLIGR---ENSIARLGIHGLYWLYHVNIP 352
Query: 573 SNLLLKGKNIIYLTQTNADTPFQGVMYDYIRLEQPP 608
S+LL+ G N I+ TQ +PFQG MYDYIRLE PP
Sbjct: 353 SSLLIDGTNTIFFTQPRCTSPFQGFMYDYIRLEGPP 460
>AW691456
Length = 660
Score = 123 bits (309), Expect = 2e-28
Identities = 61/114 (53%), Positives = 81/114 (70%)
Frame = +2
Query: 1 MARPEGYILGISYNGIDNVLDSENQELDRGYFDVVWNEPGEQNLSKSTFQRIHGTNFSVI 60
+A P GY+LGISY G++NVL++ N+ DRGYFDVV+ + TFQR+HGT+FSV+
Sbjct: 341 LANPGGYVLGISYAGMENVLEAGNEADDRGYFDVVFGK---------TFQRVHGTSFSVV 493
Query: 61 AADENMVEVSFSRLWTHSMSGKNVPINIDIRYIFRSGDSGFYSYAIFNRPKGMP 114
+E++VEVSF R W+ SM +VPINID RYI R GDS FY+Y +F K +P
Sbjct: 494 TQNEHIVEVSFLRAWSSSMGVSSVPINIDQRYIVRKGDS*FYTYVVF*TTKRIP 655
>AL377340 similar to GP|2160178|gb| F21M12.27 gene product {Arabidopsis
thaliana}, partial (22%)
Length = 453
Score = 115 bits (289), Expect = 4e-26
Identities = 68/147 (46%), Positives = 90/147 (60%), Gaps = 11/147 (7%)
Frame = +1
Query: 131 RFKYMAISDTRQRNMPAMKDRNT--GQVLAYPEAVLLTNPINPQFRGEVDDKYQYSCENK 188
RF YMA +D RQRNMP DR GQ LAYPEAVLL NPI P+ +GEVDDKYQYSC+NK
Sbjct: 19 RFHYMARADKRQRNMPLPDDRVAPRGQALAYPEAVLLVNPIEPELKGEVDDKYQYSCDNK 198
Query: 189 DTQF-----MDGLVLILI----HQWVSG**HRVMSFAMVDPLSKILLLMLDQSLFRTHYA 239
D+Q MD V + +++ SG + + V P + + L HY+
Sbjct: 199 DSQVHGWICMDPAVGFWLITPSNEFRSGGPVKQNLTSHVGPTTLAVFLS-------AHYS 357
Query: 240 GKEVTMAFQEGETYKKVFGPVFVYLNT 266
G+++ F+ GE +KKVFGPVF+Y+N+
Sbjct: 358 GEDLVPKFKAGEAWKKVFGPVFIYVNS 438
>BF641076 similar to GP|2160180|gb|A F21M12.30 gene product {Arabidopsis
thaliana}, partial (8%)
Length = 613
Score = 59.7 bits (143), Expect = 3e-09
Identities = 30/75 (40%), Positives = 44/75 (58%)
Frame = +1
Query: 1 MARPEGYILGISYNGIDNVLDSENQELDRGYFDVVWNEPGEQNLSKSTFQRIHGTNFSVI 60
++ P+G + GI YNG+DN+L+ N+E +RGY+D+VW+ PG SK F T F
Sbjct: 343 LSNPDGIVTGIRYNGVDNLLEVLNKETNRGYWDLVWSAPG----SKGIFDVYANTTFFCY 510
Query: 61 AADENMVEVSFSRLW 75
DE SF+ +W
Sbjct: 511 YLDE-----SFAIIW 540
>AW690576 similar to GP|16323222|gb| AT4g24430/T22A6_260 {Arabidopsis
thaliana}, partial (7%)
Length = 413
Score = 58.2 bits (139), Expect = 9e-09
Identities = 24/39 (61%), Positives = 30/39 (76%)
Frame = +2
Query: 412 RNGPTIWEIGIPDRLTSEFHVPEPYPTLMNKLYTEGRDK 450
R+GPT+WEIGIPDR +EF+VP+P P +NKLY DK
Sbjct: 2 RDGPTLWEIGIPDRSAAEFYVPDPNPKYINKLYVNHPDK 118
>TC83396 similar to GP|22858664|gb|AAN05792.1 unknown {Gossypium hirsutum},
partial (40%)
Length = 921
Score = 28.9 bits (63), Expect = 6.0
Identities = 17/70 (24%), Positives = 27/70 (38%)
Frame = +3
Query: 226 LLMLDQSLFRTHYAGKEVTMAFQEGETYKKVFGPVFVYLNTASSENDNATLWSDAVQQLS 285
L+ + +S++ H K + + YLNT S N + + + QL
Sbjct: 303 LMCMWRSMYECHQVQKHIVQQLE--------------YLNTIPSNNPTSEIHRQSTLQLE 440
Query: 286 KEVQSWPYDF 295
EVQ W F
Sbjct: 441 LEVQQWHQSF 470
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.140 0.436
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,507,447
Number of Sequences: 36976
Number of extensions: 284267
Number of successful extensions: 1084
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 1068
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1074
length of query: 609
length of database: 9,014,727
effective HSP length: 102
effective length of query: 507
effective length of database: 5,243,175
effective search space: 2658289725
effective search space used: 2658289725
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146709.12


