

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146709.1 - phase: 0 /pseudo
(495 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
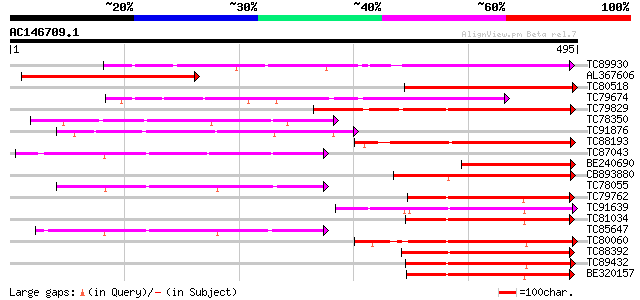
Score E
Sequences producing significant alignments: (bits) Value
TC89930 similar to PIR|T47694|T47694 probable serine/threonine-s... 260 1e-69
AL367606 weakly similar to GP|20514794|gb| Hypothetical protein ... 233 1e-61
TC80518 weakly similar to PIR|T49986|T49986 lectin-like protein ... 204 8e-53
TC79674 similar to GP|5545339|dbj|BAA82556.1 lectin-like protein... 187 6e-48
TC79829 similar to GP|20146232|dbj|BAB89014. similar to receptor... 167 9e-42
TC78350 similar to GP|1755066|gb|AAB51442.1| lectin precursor {S... 164 6e-41
TC91876 weakly similar to GP|1771455|emb|CAA93830.1 lectin 4 {Ph... 161 6e-40
TC88193 similar to PIR|G96602|G96602 probable receptor protein k... 160 1e-39
TC87043 weakly similar to GP|1755066|gb|AAB51442.1| lectin precu... 158 4e-39
BE240690 similar to PIR|T49986|T49 lectin-like protein kinase-li... 156 2e-38
CB893880 similar to PIR|T51453|T51 serine/threonine specific pro... 155 3e-38
TC78055 weakly similar to GP|1755066|gb|AAB51442.1| lectin precu... 154 1e-37
TC79762 similar to PIR|H96731|H96731 hypothetical protein F5A18.... 151 5e-37
TC91639 similar to PIR|T05181|T05181 S-receptor kinase (EC 2.7.1... 150 1e-36
TC81034 similar to GP|20453162|gb|AAM19822.1 At5g56885 {Arabidop... 150 1e-36
TC85647 similar to GP|6539559|dbj|BAA88176.1 ESTs AU068544(C3043... 148 4e-36
TC80060 similar to PIR|D96574|D96574 hypothetical protein T3F20.... 148 5e-36
TC88392 similar to GP|11275527|dbj|BAB18292. putative receptor-l... 147 7e-36
TC89432 weakly similar to GP|9294592|dbj|BAB02873.1 protein kina... 146 2e-35
BE320157 weakly similar to PIR|T04835|T04 probable serine/threon... 145 5e-35
>TC89930 similar to PIR|T47694|T47694 probable serine/threonine-specific
protein kinase - Arabidopsis thaliana, partial (53%)
Length = 1449
Score = 260 bits (664), Expect = 1e-69
Identities = 166/433 (38%), Positives = 241/433 (55%), Gaps = 22/433 (5%)
Frame = +2
Query: 83 VGRVFYSEKVLLWDSNTGKLTDFTTHYTFVINTQGRSPSLYGHGLAFFLVPYGFEIPLNS 142
+G FY + + + TGK+ F++ +F + P L GHG+AF +VP L S
Sbjct: 38 LGHAFYPQPFQIKNKTTGKVFSFSS--SFALACVPEYPKLGGHGMAFTIVPSKDLKALPS 211
Query: 143 DGGFMGLFNTTTMVSSSNQIVHVEFDSFANREFREAR-GHVGININSIISSATTP----- 196
++GL N++ + + SN + VEFD+ + EF + HVGI+INS+ S+AT
Sbjct: 212 Q--YLGLLNSSDVGNFSNHLFAVEFDTVQDFEFGDINDNHVGIDINSMRSNATITAGYYS 385
Query: 197 -----WNASKHSGDTAEVWIRYNSTTKNLTVSWKYQTTSNPQENTSLSISIDLMKVMPEW 251
N S G VW+ Y+S+ + ++V+ TSN + L+ +DL + +
Sbjct: 386 DDDMVHNISIKGGKPILVWVDYDSSLELISVT--LSPTSNKPKKPILTFHMDLSPLFLDT 559
Query: 252 ITVGFSAATSYVQELNYLLSWEFN----------STLATSGDSKTKETRLVIILTVSCGV 301
+ VGFSA+T + +Y+L W F S L K K T L++ L++ +
Sbjct: 560 MYVGFSASTGLLASSHYVLGWSFKINGPAPFLDLSKLPKLPHPKKKHTSLILGLSLGSAL 739
Query: 302 IVIGVGALVAYALFWRKRKRSNKQKEEAMHLTSMNDDLERGAGPRRFTYKELDLATNNFS 361
IV+ +A+ F+ RK N EA L GP R+TY+EL AT F
Sbjct: 740 IVL---CSMAFG-FYIYRKIKNADVIEAWELE---------VGPHRYTYQELKKATKGFK 880
Query: 362 KDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLL 421
+ + LGQGGFG VY G +QVAVK++S S+QG +E+V+E+ I +LRHRNLV LL
Sbjct: 881 EKQLLGQGGFGKVYNGILPKSKIQVAVKRVSHESKQGLREFVSEIASIGRLRHRNLVMLL 1060
Query: 422 GWCHDKGEFLLVYEFMPNGSLDSHLF-GKRTPLSWGVRHKIALGLASGLLYLHEEWERCV 480
GWC +G+ LLVY++M NGSLD +LF LSW R KI G+ASGLLYLHE +E+ V
Sbjct: 1061GWCRRRGDLLLVYDYMANGSLDKYLFEDSEYVLSWEQRFKIIKGVASGLLYLHEGYEQVV 1240
Query: 481 VHRDIKSSNVMLD 493
+HRD+K+SNV+LD
Sbjct: 1241IHRDVKASNVLLD 1279
>AL367606 weakly similar to GP|20514794|gb| Hypothetical protein {Oryza
sativa (japonica cultivar-group)}, partial (8%)
Length = 548
Score = 233 bits (595), Expect = 1e-61
Identities = 113/156 (72%), Positives = 129/156 (82%), Gaps = 1/156 (0%)
Frame = -3
Query: 11 LIVSVE-TQSMASSLYILTLFYMSLIFFNIPASSIHFKYPSFDPSDANIVYQGSAAPRDG 69
L++S++ TQ MA Y L LF++SLI A SIHF+ PSF+P+DANI+YQGSAAP DG
Sbjct: 468 LLISLKITQGMAPCFYTLPLFFLSLILLTFHAYSIHFQIPSFNPNDANIIYQGSAAPVDG 289
Query: 70 EVNFNINENYSCQVGRVFYSEKVLLWDSNTGKLTDFTTHYTFVINTQGRSPSLYGHGLAF 129
EV+FNIN NYSCQVG YS+KVLLWDS TG+LTDFTTHYTF+INT+GRSPS YGHGLAF
Sbjct: 288 EVDFNINGNYSCQVGWALYSKKVLLWDSKTGQLTDFTTHYTFIINTRGRSPSFYGHGLAF 109
Query: 130 FLVPYGFEIPLNSDGGFMGLFNTTTMVSSSNQIVHV 165
FL YGFEIP NS GG MGLFNTTTM+SSSN IVHV
Sbjct: 108 FLAAYGFEIPPNSSGGLMGLFNTTTMLSSSNHIVHV 1
>TC80518 weakly similar to PIR|T49986|T49986 lectin-like protein kinase-like
- Arabidopsis thaliana, partial (34%)
Length = 1260
Score = 204 bits (518), Expect = 8e-53
Identities = 97/151 (64%), Positives = 120/151 (79%)
Frame = +1
Query: 345 PRRFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVT 404
P++F Y+EL +TNNF+ + K+G GGFGAVYKG+ DL VA+KK+S+ S QG KEY +
Sbjct: 10 PKKFIYEELARSTNNFANEHKIGAGGFGAVYKGFIRDLKHHVAIKKVSKESNQGVKEYAS 189
Query: 405 EVKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFGKRTPLSWGVRHKIALG 464
EVKVISQLRH+NLV+L GWCH + + LLVYEF+ NGSLDS++F + L W VR+ IA G
Sbjct: 190 EVKVISQLRHKNLVQLYGWCHKQNDLLLVYEFVENGSLDSYIFKGKGLLIWTVRYNIARG 369
Query: 465 LASGLLYLHEEWERCVVHRDIKSSNVMLDSS 495
LAS LLYLHEE E CV+HRDIKSSNVMLDS+
Sbjct: 370 LASALLYLHEECEHCVLHRDIKSSNVMLDSN 462
>TC79674 similar to GP|5545339|dbj|BAA82556.1 lectin-like protein kinase
{Populus nigra}, partial (29%)
Length = 1580
Score = 187 bits (476), Expect = 6e-48
Identities = 125/363 (34%), Positives = 193/363 (52%), Gaps = 10/363 (2%)
Frame = +2
Query: 84 GRVFYSEKVLLWD--SNTGKLTDFTTHYTFVINTQGRSPSLYGHGLAFFLVPYGFEIPLN 141
GRVFY E LWD S+TGKL F T +F+IN + G G+AF + P IPLN
Sbjct: 278 GRVFYKEPFKLWDGSSSTGKLVSFNT--SFLINIYRYNNGTPGEGIAFIIAP-SLSIPLN 448
Query: 142 SDGGFMGLFNTTTMVSSSNQIVHVEFDSFANREFREARGHVGININSIISSATTPWNASK 201
S GG++GL N TT + +N+ V VE D+ ++F + + H+G++INS+ S+ + P +
Sbjct: 449 SSGGYLGLTNATTDGNVNNRFVAVELDT-VKQDFDDDKNHIGLDINSVRSNVSVPLDLEL 625
Query: 202 HSGDTA--EVWIRYNSTTKNLTVSWKYQTTSN-----PQENTSLSISIDLMKVMPEWITV 254
T +W+ Y+ KNL++ Q + + +S +DL +V+ +
Sbjct: 626 SPIGTRFHVLWVEYDGDRKNLSIYMAEQPSQDLPIVKKPAKPIISSVLDLRQVVSQNSYF 805
Query: 255 GFSAATSYVQELNYLLSWEFNSTLATSGDSKTKETRLVIILTVSCGVIVIGVGALVAYAL 314
GFSA+T ELN +L W + + D+ E L I L V V+V+ ++A
Sbjct: 806 GFSASTGITVELNCVLRWNISMEVF---DNNKNEKNLSIGLGVGIPVLVL----ILAGGG 964
Query: 315 FWRKRKRSNKQKEEAMHLTSMNDDLERGAG-PRRFTYKELDLATNNFSKDRKLGQGGFGA 373
FW K+ +E + + L+ G PR F+++EL ATNNF + KLGQGG+G
Sbjct: 965 FWYYYLCKKKKSDEHGSTSQIMGTLKSLPGTPREFSFQELKKATNNFDEKHKLGQGGYGV 1144
Query: 374 VYKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLV 433
VY+G L+VAVK SR + +++ E+ +I++LRH++LVKL G H L+
Sbjct: 1145VYRGTLPKEKLEVAVKMFSRDKMKSTDDFLAELTIINRLRHKHLVKLQGQFHFSFS*LIF 1324
Query: 434 YEF 436
+F
Sbjct: 1325IKF 1333
>TC79829 similar to GP|20146232|dbj|BAB89014. similar to receptor protein
kinase {Oryza sativa (japonica cultivar-group)}, partial
(40%)
Length = 1736
Score = 167 bits (423), Expect = 9e-42
Identities = 92/231 (39%), Positives = 144/231 (61%), Gaps = 2/231 (0%)
Frame = +2
Query: 266 LNYLLSWEFNSTLATSGDSKTKETRLVIIL-TVSCGVIVIGVGALVAYALFWRKRKRSNK 324
+N+ + N+T +S + + I+L ++C V + + L L R + +
Sbjct: 122 INFNMGLYQNATSTSSKSGISTGAIVGIVLGAIACAVTLSAIVTL----LILRTKLKDYH 289
Query: 325 QKEEAMHLTSMNDDLERGAGPRRFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDL 384
+ H++ + ++ G R FTY+EL ATNNFS ++GQGG+G VYKG +
Sbjct: 290 AVSKRRHVSKIKIKMD---GVRSFTYEELSSATNNFSSSAQVGQGGYGKVYKGVISG-GT 457
Query: 385 QVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDS 444
VA+K+ GS QG+KE++TE+ ++S+L HRNLV L+G+C ++GE +LVYE+MPNG+L
Sbjct: 458 AVAIKRAQEGSLQGEKEFLTEISLLSRLHHRNLVSLIGYCDEEGEQMLVYEYMPNGTLRD 637
Query: 445 HL-FGKRTPLSWGVRHKIALGLASGLLYLHEEWERCVVHRDIKSSNVMLDS 494
HL + PL++ +R KIALG A GL+YLH E + + HRD+K+SN++LDS
Sbjct: 638 HLSVSAKEPLTFIMRLKIALGSAKGLMYLHNEADPPIFHRDVKASNILLDS 790
>TC78350 similar to GP|1755066|gb|AAB51442.1| lectin precursor {Sophora
japonica}, partial (33%)
Length = 1032
Score = 164 bits (416), Expect = 6e-41
Identities = 106/281 (37%), Positives = 154/281 (54%), Gaps = 12/281 (4%)
Frame = +1
Query: 19 SMASSLYILTLFYMSLIFFNIPASSIH---FKYPSFDPSDANIVYQGSAAPRDGEVNF-N 74
+MA + + ++++I F I A +++ F +FDP NI +G+A DG ++ N
Sbjct: 25 TMAINTSRTQILFITIISFLILAQNVNSAAFTVSNFDPYKTNIELEGNAFISDGSIHLTN 204
Query: 75 INENYSCQVGRVFYSEKVLLWDSNTGKLTDFTTHYTFVINTQGRSPSLYGHGLAFFLVPY 134
+ N GR + V LWD++TG L FT+ ++F + G P L G G+ FF+ P+
Sbjct: 205 VIPN---SAGRASWGGPVRLWDADTGNLAGFTSVFSFEVAPAG--PGLIGDGITFFIAPF 369
Query: 135 GFEIPLNSDGGFMGLFNTTTMVSS-SNQIVHVEFDSFANRE----FREARGHVGININSI 189
IP NS GGF+GLFN T +++ N+IV VEFDSF + A HVGI++NSI
Sbjct: 370 NSHIPKNSSGGFLGLFNAETALNTYQNRIVAVEFDSFGGNSGGNPWDPAYPHVGIDVNSI 549
Query: 190 ISSATTPWNA-SKHSGDTAEVWIRYNSTTKNLTVSWKYQTTSNPQENTSLSIS--IDLMK 246
S T PW S +G A ++ Y KNL+V +Y N TS S+S IDL
Sbjct: 550 ASVTTAPWKTGSILTGFNAIAFVNYEPVEKNLSVVVRYPG-GNFVNGTSNSVSFIIDLRT 726
Query: 247 VMPEWITVGFSAATSYVQELNYLLSWEFNSTLATSGDSKTK 287
V+PEW+ +GFS AT + EL+ +LSW F S+ G T+
Sbjct: 727 VLPEWVRIGFSGATGQLVELHKILSWTFKSSFQ*CGYKLTR 849
>TC91876 weakly similar to GP|1771455|emb|CAA93830.1 lectin 4 {Phaseolus
lunatus}, partial (20%)
Length = 876
Score = 161 bits (407), Expect = 6e-40
Identities = 101/279 (36%), Positives = 149/279 (53%), Gaps = 16/279 (5%)
Frame = +3
Query: 42 SSIHFKYPSFDPSD--ANIVYQGSAAPRDGEVNFNINENYSCQVGRVFYSEKVLLWDSNT 99
+S+ F +FD +N+ YQG +G ++ N +Y +VGR FYS+ + LWD T
Sbjct: 36 NSLLFNITNFDDPTVASNMSYQGDGKSTNGSIDLN-KVSYLFRVGRAFYSQPLHLWDKKT 212
Query: 100 GKLTDFTTHYTFVINTQGRSPSLYGHGLAFFLVPYGFEIPLNSDGGFMGLFNTTTMVSS- 158
LT FTT ++F I+ + + YG G F+L P G++IP NS GG GLFN TT +
Sbjct: 213 NTLTSFTTRFSFTIDKL--NDTTYGDGFVFYLAPLGYQIPPNSAGGVYGLFNATTNSNFV 386
Query: 159 SNQIVHVEFDSFANREFREARGHVGININSIISSATTPWNASKHSGDTAEVWIRYNSTTK 218
N +V VEFD+F + HVGI+ N++ S A ++ K+ G V I YNS K
Sbjct: 387 MNYVVGVEFDTFVGPTDPPMK-HVGIDDNALTSVAFGKFDIDKNLGRVCYVLIDYNSDEK 563
Query: 219 NLTVSWKYQTT----SNPQENTSLSISIDLMKVMPEWITVGFSAATSYVQELNYLLSWEF 274
L V W ++ N+S+S IDLMK +PE++ +GFSA+T E N + SWEF
Sbjct: 564 MLEVFWSFKGRFVKGDGKYGNSSISYQIDLMKKLPEFVNIGFSASTGLSTESNVIHSWEF 743
Query: 275 NSTLATS---------GDSKTKETRLVIILTVSCGVIVI 304
+S L S D K +++++ V VI++
Sbjct: 744 SSNLEDSNSTTSLVEGNDGKGSLKTVIVVVAVIVPVILV 860
>TC88193 similar to PIR|G96602|G96602 probable receptor protein kinase
F14G9.24 [imported] - Arabidopsis thaliana, partial
(20%)
Length = 1815
Score = 160 bits (405), Expect = 1e-39
Identities = 90/197 (45%), Positives = 126/197 (63%), Gaps = 4/197 (2%)
Frame = +3
Query: 302 IVIGVGA---LVAYALFWRKRKRSNKQKEEAMHLTSMNDDLER-GAGPRRFTYKELDLAT 357
+V+GVGA L ++F+ R+R L + +DDL P F+Y EL AT
Sbjct: 417 VVVGVGAVCFLAVSSIFYILRRRK---------LYNDDDDLVGIDTMPNTFSYYELKNAT 569
Query: 358 NNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNL 417
++F++D KLG+GGFG VYKG D VAVK++S GS QGK +++ E+ IS ++HRNL
Sbjct: 570 SDFNRDNKLGEGGFGPVYKGTLNDGRF-VAVKQLSIGSHQGKSQFIAEIATISAVQHRNL 746
Query: 418 VKLLGWCHDKGEFLLVYEFMPNGSLDSHLFGKRTPLSWGVRHKIALGLASGLLYLHEEWE 477
VKL G C + + LLVYE++ N SLD LFG L+W R+ + +G+A GL YLHEE
Sbjct: 747 VKLYGCCIEGNKRLLVYEYLENKSLDQALFGNVLFLNWSTRYDVCMGVARGLTYLHEESR 926
Query: 478 RCVVHRDIKSSNVMLDS 494
+VHRD+K+SN++LDS
Sbjct: 927 LRIVHRDVKASNILLDS 977
>TC87043 weakly similar to GP|1755066|gb|AAB51442.1| lectin precursor
{Sophora japonica}, partial (28%)
Length = 1029
Score = 158 bits (400), Expect = 4e-39
Identities = 99/280 (35%), Positives = 153/280 (54%), Gaps = 7/280 (2%)
Frame = +1
Query: 6 SKLSKLIVSVETQSMASSLYILTLFYMSLIFFNIPASSIHFKYPSFDPSDANIVYQGSAA 65
+ + KL+ + S++ S++ F++ L+ N+ + S+ F +P FD NIV G A
Sbjct: 37 NSIPKLLATQNPFSVSLSIF----FFLLLLINNVKSDSLSFNFPKFDTDALNIVIDGDAK 204
Query: 66 PRDGEVNFNINENYSC----QVGRVFYSEKVLLWDSNTGKLTDFTTHYTFVINTQGRSPS 121
G + + + VG + + L D +GK+ DFTT ++FV+N +G
Sbjct: 205 TTGGVLQLTKKDQFGNPSPHSVGFSAFFGAIQLSDKQSGKVADFTTEFSFVVNPKGSQ-- 378
Query: 122 LYGHGLAFFLVPYGFEIP-LNSDGGFMGLFNTTTMVSSS-NQIVHVEFDSFANREFREAR 179
L+G G AF++ ++ P +SDGGF+GLF+ T ++S N IV VEFDSFAN E+
Sbjct: 379 LHGDGFAFYIASLDYDFPEKSSDGGFLGLFDKETAFNTSKNSIVAVEFDSFAN-EWDPNS 555
Query: 180 GHVGININSIISSATTPWNASKH-SGDTAEVWIRYNSTTKNLTVSWKYQTTSNPQENTSL 238
H+GI+IN+I SS + PW + G + I YN+ +K+L+V Y S + + +
Sbjct: 556 PHIGIDINTIESSISVPWPIDRQPQGTIGKARISYNTASKDLSVFVTYPN-SPAKVDVIV 732
Query: 239 SISIDLMKVMPEWITVGFSAATSYVQELNYLLSWEFNSTL 278
S ID V+ EW+ VGFS AT V E + +LSW F S L
Sbjct: 733 SYPIDFASVLSEWVYVGFSGATGQVAETHDILSWSFVSNL 852
>BE240690 similar to PIR|T49986|T49 lectin-like protein kinase-like -
Arabidopsis thaliana, partial (25%)
Length = 634
Score = 156 bits (394), Expect = 2e-38
Identities = 71/100 (71%), Positives = 86/100 (86%)
Frame = +2
Query: 395 SRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFGKRTPLS 454
S QG KE+ +EV +IS+LRHRNLV+L+GWCH + + LLVYE+MPNGSLD HLF K++ L
Sbjct: 2 SHQGIKEFASEVTIISKLRHRNLVQLIGWCHQRKKLLLVYEYMPNGSLDIHLFKKQSLLK 181
Query: 455 WGVRHKIALGLASGLLYLHEEWERCVVHRDIKSSNVMLDS 494
WGVR+ IA GLAS LLYLHEEWE+CVVHRDIK+SN+MLDS
Sbjct: 182 WGVRYTIAKGLASALLYLHEEWEQCVVHRDIKASNIMLDS 301
>CB893880 similar to PIR|T51453|T51 serine/threonine specific protein
kinase-like - Arabidopsis thaliana, partial (42%)
Length = 645
Score = 155 bits (392), Expect = 3e-38
Identities = 74/168 (44%), Positives = 113/168 (67%), Gaps = 9/168 (5%)
Frame = +3
Query: 336 NDDLERGAGPRRFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFAD---------LDLQV 386
+++L+ + R+FT+ EL +AT NF + LG+GGFG V+KG+ + L V
Sbjct: 27 SEELKVSSDLRKFTFNELKMATRNFRPESLLGEGGFGCVFKGWIEENGTAPVKPGTGLTV 206
Query: 387 AVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHL 446
AVK ++ QG KE++ E+ ++ + H NLVKL+G+C + + LLVY+FMP GSL++HL
Sbjct: 207 AVKTLNHDGLQGHKEWLAELNILGDIVHPNLVKLIGFCIEDDQRLLVYQFMPRGSLENHL 386
Query: 447 FGKRTPLSWGVRHKIALGLASGLLYLHEEWERCVVHRDIKSSNVMLDS 494
F + PL W +R KIALG A GL +LHEE +R +++RD K+SN++LD+
Sbjct: 387 FRRSLPLPWSIRMKIALGAAKGLNFLHEEAQRPIIYRDFKTSNILLDA 530
>TC78055 weakly similar to GP|1755066|gb|AAB51442.1| lectin precursor
{Sophora japonica}, partial (24%)
Length = 1060
Score = 154 bits (388), Expect = 1e-37
Identities = 89/247 (36%), Positives = 135/247 (54%), Gaps = 10/247 (4%)
Frame = +1
Query: 42 SSIHFKYPSFDPSDANIVYQGSAAPRDGEVNFNINENYSCQ-----VGRVFYSEKVLLWD 96
S+I F FD NI +G + +G ++ + YS + VG + + +WD
Sbjct: 118 STISFSITKFDDESPNIFVKGDTSISNGVLSLTKTDKYSGKPLQKSVGHATHLTPIHIWD 297
Query: 97 SNTGKLTDFTTHYTFVINTQGRSPSLYGHGLAFFLVPYGFEIPLNSDGGFMGLFNT-TTM 155
+G+L DF+T ++F++NT G S L+G G FFL P F++P NS GG++GLFN T +
Sbjct: 298 ETSGELADFSTSFSFIVNTNGSS--LHGDGFTFFLGPLHFDLPKNSSGGYLGLFNPETAL 471
Query: 156 VSSSNQIVHVEFDSFANREFREARG---HVGININSIISSATTPWNASKHSGDT-AEVWI 211
+ S N IV +EFDSF N + H+GI++ SI S +T W + + E I
Sbjct: 472 IPSQNPIVAIEFDSFTNGWDPASPSQYTHIGIDVGSIDSVSTADWPLNVLPRNALGEARI 651
Query: 212 RYNSTTKNLTVSWKYQTTSNPQENTSLSISIDLMKVMPEWITVGFSAATSYVQELNYLLS 271
YNS +K L+ Y E+T +S +DL V+PEW+ VGFSAAT + E + +++
Sbjct: 652 NYNSESKRLSAFVDYPGLG---ESTGVSFVVDLRSVLPEWVRVGFSAATGELVETHDIIN 822
Query: 272 WEFNSTL 278
W F + L
Sbjct: 823 WSFETAL 843
>TC79762 similar to PIR|H96731|H96731 hypothetical protein F5A18.8
[imported] - Arabidopsis thaliana, partial (55%)
Length = 1564
Score = 151 bits (382), Expect = 5e-37
Identities = 76/149 (51%), Positives = 106/149 (71%), Gaps = 3/149 (2%)
Frame = +3
Query: 348 FTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVTEVK 407
F+Y+ L AT NF+ KLG+GGFG VYKG +D +VAVKK+S+ S QGKKE++ E K
Sbjct: 348 FSYETLLSATKNFNATHKLGEGGFGPVYKGKLSD-GREVAVKKLSQTSNQGKKEFMNEAK 524
Query: 408 VISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLF---GKRTPLSWGVRHKIALG 464
++++++H+N+V LLG+C E +LVYE++P+ SLD LF KR L W R I G
Sbjct: 525 LLARVQHKNVVNLLGYCVHGTEKILVYEYVPHESLDKFLFKEAEKREQLDWKRRFGIITG 704
Query: 465 LASGLLYLHEEWERCVVHRDIKSSNVMLD 493
+A GLLYLHE+ C++HRDIK+SN++LD
Sbjct: 705 VAKGLLYLHEDSHNCIIHRDIKASNILLD 791
>TC91639 similar to PIR|T05181|T05181 S-receptor kinase (EC 2.7.1.-)
T6K22.120 precursor - Arabidopsis thaliana, partial
(28%)
Length = 1313
Score = 150 bits (379), Expect = 1e-36
Identities = 92/231 (39%), Positives = 136/231 (58%), Gaps = 20/231 (8%)
Frame = +3
Query: 285 KTKETRLVIILTVSCGVIVIGVGALVAYALFWRKRKRSNKQKEEAMHLTSMNDDLERGA- 343
K ++ +VIILT G+I IG+ L+ + + R+ K S + + + + + E A
Sbjct: 132 KNEKIMMVIILTSLAGLICIGIIVLLVWR-YKRQLKASCSKNSDVLPVFDAHKSREMSAE 308
Query: 344 ----------GPRR-------FTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQV 386
G + F + + ATNNFS++ KLGQGGFG VYKG + ++
Sbjct: 309 IPGSVELGLEGNQLSKVELPFFNFSCMSSATNNFSEENKLGQGGFGPVYKGKLPSGE-EI 485
Query: 387 AVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHL 446
AVK++SR S QG E+ E+++ +QL+HRNLVKL+G + E LLVYEFM N SLD L
Sbjct: 486 AVKRLSRRSGQGLDEFKNEMRLFAQLQHRNLVKLMGCSIEGDEKLLVYEFMLNKSLDRFL 665
Query: 447 FG--KRTPLSWGVRHKIALGLASGLLYLHEEWERCVVHRDIKSSNVMLDSS 495
F K+T L W R++I G+A GLLYLH + ++HRD+K+SN++LD +
Sbjct: 666 FDPIKKTQLDWARRYEIIEGIARGLLYLHRDSRLRIIHRDLKASNILLDEN 818
>TC81034 similar to GP|20453162|gb|AAM19822.1 At5g56885 {Arabidopsis
thaliana}, partial (35%)
Length = 1645
Score = 150 bits (379), Expect = 1e-36
Identities = 76/152 (50%), Positives = 104/152 (68%), Gaps = 4/152 (2%)
Frame = +2
Query: 346 RRFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVTE 405
R F+ +++ AT+NF R LG+GGFG VY G D +VAVK + QG +E++ E
Sbjct: 932 RTFSMDDIEKATDNFHASRILGEGGFGLVYSGVLGD-GTKVAVKVLKSKDHQGDREFLAE 1108
Query: 406 VKVISQLRHRNLVKLLGWCHDKGEF-LLVYEFMPNGSLDSHLFG---KRTPLSWGVRHKI 461
V+++S+L HRNL+KL+G C ++ F LVYE +PNGSL+SHL G ++ L WG R KI
Sbjct: 1109 VEMLSRLHHRNLIKLIGICAEEDSFRCLVYELIPNGSLESHLHGVEWEKRALDWGARMKI 1288
Query: 462 ALGLASGLLYLHEEWERCVVHRDIKSSNVMLD 493
ALG A GL YLHE+ CV+HRD KSSN++L+
Sbjct: 1289 ALGAARGLSYLHEDSSPCVIHRDFKSSNILLE 1384
>TC85647 similar to GP|6539559|dbj|BAA88176.1 ESTs AU068544(C30430)
C98487(E0325) D23445(C2825) AU068543(C30430)
C71774(E0325) D22458(C11152), partial (11%)
Length = 1366
Score = 148 bits (374), Expect = 4e-36
Identities = 94/267 (35%), Positives = 142/267 (52%), Gaps = 11/267 (4%)
Frame = +2
Query: 23 SLYILTLFYMSLIFFNIPASSIHFKYPSFDPSDANIVYQGSAAPRDGEVNFNINENYSC- 81
S+++LT L+ N+ + S F +P FD +I+ G A +G + +
Sbjct: 395 SVFLLTFL---LLITNVKSDSFSFNFPKFDTDTKSIIIDGDANTTNGVLQLTKKDQLGNP 565
Query: 82 ---QVGRVFYSEKVLLWDSNTGKLTDFTTHYTFVINTQGRSPSLYGHGLAFFLVPYGFEI 138
G F+ + L D +GK+ DFTT ++FV+N +G L+G G FF+ +E
Sbjct: 566 SPHSFGLSFFLGAIHLSDKQSGKVADFTTEFSFVVNPKGSQ--LHGDGFTFFIASLDYEF 739
Query: 139 P-LNSDGGFMGLFNTTTMVSSS-NQIVHVEFDSFANREFREARG---HVGININSIISSA 193
P +SDGGF+GLF+ + ++S N IV VEFDSF N + G H+GI+IN+I SSA
Sbjct: 740 PEKSSDGGFLGLFDKESAFNTSQNSIVAVEFDSFRNEWDPQIAGNSPHIGIDINTIRSSA 919
Query: 194 TTPWNASK-HSGDTAEVWIRYNSTTKNLTVSWKYQTTSNPQ-ENTSLSISIDLMKVMPEW 251
T W + G + I YN +K LT Y + P E T++S ++D ++PE+
Sbjct: 920 TALWPIDRVPEGSIGKAHISYNPASKKLTALVTY--LNGPVIEETAVSYTVDFAAILPEY 1093
Query: 252 ITVGFSAATSYVQELNYLLSWEFNSTL 278
+ VGFS AT + E + +LSW F S L
Sbjct: 1094VLVGFSGATGELAETHDILSWSFTSNL 1174
>TC80060 similar to PIR|D96574|D96574 hypothetical protein T3F20.24
[imported] - Arabidopsis thaliana, partial (32%)
Length = 1547
Score = 148 bits (373), Expect = 5e-36
Identities = 88/199 (44%), Positives = 121/199 (60%), Gaps = 5/199 (2%)
Frame = +1
Query: 302 IVIGVGALVAYALF--WRKRKRSNKQKEEAMHLTSMNDDLERGAGPRRFTYKELDLATNN 359
IVIG A V LF W+ K + + L +L+ G ++ +++ +ATNN
Sbjct: 40 IVIGSLAFVMLILFVLWKMGYLCGKDQTDKELL-----ELKTGY----YSLRQIKVATNN 192
Query: 360 FSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVK 419
F K+G+GGFG VYKG +D +AVK++S S+QG +E+V E+ +IS L+H NLVK
Sbjct: 193 FDPKNKIGEGGFGPVYKGVLSD-GAVIAVKQLSSKSKQGNREFVNEIGMISALQHPNLVK 369
Query: 420 LLGWCHDKGEFLLVYEFMPNGSLDSHLFGK---RTPLSWGVRHKIALGLASGLLYLHEEW 476
L G C + + LLVYE+M N SL LFGK R L W R KI +G+A GL YLHEE
Sbjct: 370 LYGCCIEGNQLLLVYEYMENNSLARALFGKPEQRLNLDWRTRMKICVGIARGLAYLHEES 549
Query: 477 ERCVVHRDIKSSNVMLDSS 495
+VHRDIK++NV+LD +
Sbjct: 550 RLKIVHRDIKATNVLLDKN 606
>TC88392 similar to GP|11275527|dbj|BAB18292. putative receptor-like protein
kinase {Oryza sativa (japonica cultivar-group)}, partial
(61%)
Length = 1017
Score = 147 bits (372), Expect = 7e-36
Identities = 74/151 (49%), Positives = 105/151 (69%)
Frame = +1
Query: 343 AGPRRFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKEY 402
+G F L LATN FS+ +LG+GGFG V+KG + + +VA+KK+S SRQG +E+
Sbjct: 166 SGNLLFELNTLQLATNFFSELNQLGRGGFGPVFKGLMPNGE-EVAIKKLSMESRQGIREF 342
Query: 403 VTEVKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFGKRTPLSWGVRHKIA 462
EV+++ +++H+NLV LLG C + E +LVYE++PN SLD LF K+ L W R +I
Sbjct: 343 TNEVRLLLRIQHKNLVTLLGCCAEGPEKMLVYEYLPNKSLDHFLFDKKRSLDWMTRFRIV 522
Query: 463 LGLASGLLYLHEEWERCVVHRDIKSSNVMLD 493
G+A GLLYLHEE ++HRDIK+SN++LD
Sbjct: 523 TGIARGLLYLHEEAPERIIHRDIKASNILLD 615
>TC89432 weakly similar to GP|9294592|dbj|BAB02873.1 protein kinase-like
protein {Arabidopsis thaliana}, partial (51%)
Length = 1272
Score = 146 bits (369), Expect = 2e-35
Identities = 77/152 (50%), Positives = 105/152 (68%), Gaps = 3/152 (1%)
Frame = +3
Query: 346 RRFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVTE 405
R FTYKEL ATN FS D KLG+GGFG+VY G +D LQ+AVKK+ + + + E+ E
Sbjct: 72 RIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSD-GLQIAVKKLKAMNSKAEMEFAVE 248
Query: 406 VKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFGK---RTPLSWGVRHKIA 462
V+V+ ++RH+NL+ L G+C + L+VY++MPN SL SHL G+ L+W R IA
Sbjct: 249 VEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSIA 428
Query: 463 LGLASGLLYLHEEWERCVVHRDIKSSNVMLDS 494
+G A G+LYLH E ++HRDIK+SNV+LDS
Sbjct: 429 IGSAEGILYLHHEVTPHIIHRDIKASNVLLDS 524
>BE320157 weakly similar to PIR|T04835|T04 probable serine/threonine-specific
protein kinase (EC 2.7.1.-) F21P8.70 - Arabidopsis
thaliana, partial (27%)
Length = 529
Score = 145 bits (365), Expect = 5e-35
Identities = 74/149 (49%), Positives = 104/149 (69%), Gaps = 2/149 (1%)
Frame = +1
Query: 347 RFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVTEV 406
+F + ++ ATN FS + ++G+GGFG VYKG +D Q+AVKK+SR S QG E+ E+
Sbjct: 19 QFKFSTIEAATNKFSSENEIGKGGFGIVYKGVLSD-GQQIAVKKLSRSSGQGSIEFQNEI 195
Query: 407 KVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFG--KRTPLSWGVRHKIALG 464
+I++L+HRNLV LLG+C ++ E +L+YE++PN SLD LF K L W R+KI G
Sbjct: 196 LLIAKLQHRNLVTLLGFCLEEREKMLIYEYVPNKSLDYFLFDSKKHRVLHWFERYKIIGG 375
Query: 465 LASGLLYLHEEWERCVVHRDIKSSNVMLD 493
+A G+LYLHE V+HRD+K SNV+LD
Sbjct: 376 IARGILYLHEYSRLKVIHRDLKPSNVLLD 462
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.134 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,338,342
Number of Sequences: 36976
Number of extensions: 217130
Number of successful extensions: 2285
Number of sequences better than 10.0: 557
Number of HSP's better than 10.0 without gapping: 1749
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1774
length of query: 495
length of database: 9,014,727
effective HSP length: 100
effective length of query: 395
effective length of database: 5,317,127
effective search space: 2100265165
effective search space used: 2100265165
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146709.1


