

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146708.4 + phase: 0
(298 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
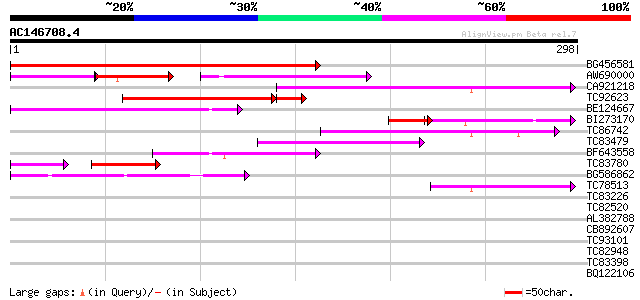
Score E
Sequences producing significant alignments: (bits) Value
BG456581 316 6e-87
AW690000 70 4e-26
CA921218 112 2e-25
TC92623 93 2e-22
BE124667 74 9e-14
BI273170 64 1e-13
TC86742 similar to PIR|T01541|T01541 hypothetical protein A_IG00... 57 9e-09
TC83479 similar to GP|23476992|emb|CAD48949. hypothetical protei... 54 7e-08
BF643558 49 3e-06
TC83780 33 2e-04
BG586862 42 2e-04
TC78513 homologue to GP|5679340|emb|CAB51773.1 hypothetical prot... 41 5e-04
TC83226 weakly similar to PIR|G86419|G86419 probable reverse tra... 40 0.001
TC82520 40 0.001
AL382788 weakly similar to GP|11322386|emb glycine receptor beta... 40 0.001
CB892607 similar to GP|8927657|gb| EST gb|N38213 comes from this... 39 0.002
TC93101 39 0.003
TC82948 38 0.004
TC83398 similar to PIR|T05150|T05150 hypothetical protein F18E5.... 36 0.016
BQ122106 similar to GP|9366656|emb|C probable similar to ring-h2... 35 0.035
>BG456581
Length = 683
Score = 316 bits (810), Expect = 6e-87
Identities = 152/163 (93%), Positives = 155/163 (94%)
Frame = +2
Query: 1 MCSFCLEQVETSDHLFLRCKFVVTLWSWLCSQLHVGLDFSSIKALMSSLPRHCSSQVRDL 60
MCSFCLEQVETSDHLFLRCKFVVTLWSWLCSQL VGLDFSS KAL+SSLPRHCSSQVRDL
Sbjct: 191 MCSFCLEQVETSDHLFLRCKFVVTLWSWLCSQLRVGLDFSSFKALLSSLPRHCSSQVRDL 370
Query: 61 YVAAVVHMVHSIWWARNNVRFSSAKVSSHAVQVRVHAFIGLSGAVSTGKCIAADAAILDA 120
YVAAVVHMVHSIWWARNNVRFSSAKVS+HAVQVRVHA IGLSGAVSTGKCIAADAAILD
Sbjct: 371 YVAAVVHMVHSIWWARNNVRFSSAKVSAHAVQVRVHALIGLSGAVSTGKCIAADAAILDV 550
Query: 121 FRIPPHRRSMREIVSVCWKPPSAPWVKVNTDGSVLNNSGACGG 163
FRIPPHRRSM E+VSVCWKPPSAPWVK NTDGS LNNSGA GG
Sbjct: 551 FRIPPHRRSMXEMVSVCWKPPSAPWVKGNTDGSXLNNSGAXGG 679
>AW690000
Length = 652
Score = 70.5 bits (171), Expect(3) = 4e-26
Identities = 35/90 (38%), Positives = 50/90 (54%)
Frame = +3
Query: 101 LSGAVSTGKCIAADAAILDAFRIPPHRRSMREIVSVCWKPPSAPWVKVNTDGSVLNNSGA 160
LS AV++ ++ IL F + H +I+ V W+PP W+K NTDGS ++S A
Sbjct: 306 LSNAVASASI--SNFLILKKFNVTIHPPKAPKIIEVIWRPPIPHWIKCNTDGSSRSHSSA 479
Query: 161 CGGLFRDHLGTFLGAFVGNLGRCSVFDTEV 190
CGG+FR+H L F N G C+ F E+
Sbjct: 480 CGGIFRNHDTDLLLCFAENTGECNAFHAEL 569
Score = 50.4 bits (119), Expect(3) = 4e-26
Identities = 21/47 (44%), Positives = 28/47 (58%)
Frame = +2
Query: 1 MCSFCLEQVETSDHLFLRCKFVVTLWSWLCSQLHVGLDFSSIKALMS 47
MCS C +Q E+S HLF C + V LW W S L+ L F S++ + S
Sbjct: 11 MCSLCCKQAESSLHLFFECSYAVNLWCWFASVLNKTLHFQSLEDMWS 151
Score = 34.7 bits (78), Expect(3) = 4e-26
Identities = 15/43 (34%), Positives = 30/43 (68%), Gaps = 2/43 (4%)
Frame = +1
Query: 46 MSSLPRHCSS--QVRDLYVAAVVHMVHSIWWARNNVRFSSAKV 86
+S RH S Q+ ++ A ++++++S+W+ARN +RFS+ K+
Sbjct: 124 LSVFGRHVVSL*QIMEVITATMINIMNSVWFARN*LRFSNKKI 252
>CA921218
Length = 707
Score = 112 bits (280), Expect = 2e-25
Identities = 58/159 (36%), Positives = 87/159 (54%), Gaps = 2/159 (1%)
Frame = -3
Query: 141 PSAPWVKVNTDGSVLNNSGACGGLFRDHLGTFLGAFVGNLGRCSVFDTEVSGFILVMEHA 200
P W+ NT+GS N+ ACGG FR+ FL F N G + ++SG + +E A
Sbjct: 594 PEPLWINCNTNGSANINTSACGGTFRNSNAYFLLCFAENTGNGNALHAKLSGAMRAIEIA 415
Query: 201 ALHGWYNIWLESDASSALMVFKNPSLVPILLRNRWHNARTL--NVQVISSHIFREGNVCV 258
A W ++WLE D+S + FK+ S++ LRNRW+N L ++ SH+FREGN C
Sbjct: 414 AARNWSHLWLELDSSLVVNAFKSKSVIHWNLRNRWNNCLFLISSMNFFVSHVFREGNQCA 235
Query: 259 DRLANLGHSVVGEVWLSTLPSDFHQVFYEDRCSMPRIRY 297
D LAN G S+ + + +P H+ + E++ P R+
Sbjct: 234 DGLANFGLSLDHLTYWNHVPPFIHRFYVENKIGWPMFRF 118
>TC92623
Length = 549
Score = 92.8 bits (229), Expect(2) = 2e-22
Identities = 41/81 (50%), Positives = 56/81 (68%)
Frame = +1
Query: 60 LYVAAVVHMVHSIWWARNNVRFSSAKVSSHAVQVRVHAFIGLSGAVSTGKCIAADAAILD 119
++ AA+VH VH+IW ARN +RFSSA VS H ++ + + LSGA S G C+ D A+L+
Sbjct: 52 VFAAALVHTVHTIWLARNAIRFSSANVSIHTSMAKISSLVALSGANSKGNCLVTDGAVLN 231
Query: 120 AFRIPPHRRSMREIVSVCWKP 140
F IPP R ++EI+SV WKP
Sbjct: 232 NFMIPPSYRRVKEIISVIWKP 294
Score = 30.4 bits (67), Expect(2) = 2e-22
Identities = 12/16 (75%), Positives = 13/16 (81%)
Frame = +3
Query: 141 PSAPWVKVNTDGSVLN 156
P+ WVK NTDGSVLN
Sbjct: 294 PTITWVKANTDGSVLN 341
>BE124667
Length = 540
Score = 73.6 bits (179), Expect = 9e-14
Identities = 39/122 (31%), Positives = 63/122 (50%)
Frame = -1
Query: 1 MCSFCLEQVETSDHLFLRCKFVVTLWSWLCSQLHVGLDFSSIKALMSSLPRHCSSQVRDL 60
MCS C ET+ HLF C F +WSWL S L+ L FS+ + S L + + Q + +
Sbjct: 387 MCSSCQASSETTFHLFFECNFATKMWSWLASLLNTPLQFSTSADMWSPLQLNWTPQCKVV 208
Query: 61 YVAAVVHMVHSIWWARNNVRFSSAKVSSHAVQVRVHAFIGLSGAVSTGKCIAADAAILDA 120
A ++++++ IW+ RNN+RF + + + + LSG + T K AA+
Sbjct: 207 ITACIINLLNVIWFRRNNIRFQDKVIDWKTAINMIISKVSLSGNL-TKKTSAANMLEFTI 31
Query: 121 FR 122
F+
Sbjct: 30 FK 25
>BI273170
Length = 391
Score = 64.3 bits (155), Expect(2) = 1e-13
Identities = 37/82 (45%), Positives = 47/82 (57%), Gaps = 3/82 (3%)
Frame = +1
Query: 219 MVFKNPSLVPILLRNRWHNA--RTLNVQVISSHIFREGNVCVDRLANLGHSV-VGEVWLS 275
M FKN S VP LRNRW N T N+ I SH+FREGN C D LAN+G S+ +WL
Sbjct: 61 MPFKNISQVPWKLRNRWENCILATRNMNFIVSHVFREGNECADMLANIGLSLNCLTIWLE 240
Query: 276 TLPSDFHQVFYEDRCSMPRIRY 297
LP +F +++ P R+
Sbjct: 241 -LPDCIKSIFIKNKLGWPNNRF 303
Score = 29.3 bits (64), Expect(2) = 1e-13
Identities = 10/23 (43%), Positives = 16/23 (69%)
Frame = +3
Query: 200 AALHGWYNIWLESDASSALMVFK 222
A LH W ++WLESD++ + F+
Sbjct: 3 AKLHNWQSLWLESDSALVVNAFQ 71
>TC86742 similar to PIR|T01541|T01541 hypothetical protein A_IG005I10.16 -
Arabidopsis thaliana, partial (19%)
Length = 2073
Score = 57.0 bits (136), Expect = 9e-09
Identities = 40/130 (30%), Positives = 58/130 (43%), Gaps = 4/130 (3%)
Frame = -1
Query: 164 LFRDHLGTFLGAFVGNLGRCSVFDTEVSGFILVMEHAALHGWYNIWLESDASSALMVFKN 223
+ RD FLGA N+G + + E ++ +E A G NI LE+D+ + F
Sbjct: 468 VIRDSQFGFLGALSCNIGHATPLEAEFCACMIAIEKAMELGLNNICLETDSLKVVNAFHK 289
Query: 224 PSLVPILLRNRWHNARTL--NVQVISSHIFREGNVCVDRLANLGH--SVVGEVWLSTLPS 279
+P +R RWHN ++ + HI REGN+ D LA G S+ W PS
Sbjct: 288 IVGIPWQMRVRWHNCIRFCHSIACVCVHIPREGNLVADALARHGQGLSLFFLQWWPAPPS 109
Query: 280 DFHQVFYEDR 289
+DR
Sbjct: 108 FIQSFLAQDR 79
>TC83479 similar to GP|23476992|emb|CAD48949. hypothetical protein {Plasmodium
falciparum 3D7}, partial (0%)
Length = 1222
Score = 53.9 bits (128), Expect = 7e-08
Identities = 31/88 (35%), Positives = 44/88 (49%)
Frame = +2
Query: 131 REIVSVCWKPPSAPWVKVNTDGSVLNNSGACGGLFRDHLGTFLGAFVGNLGRCSVFDTEV 190
R I+ WK P W K+NTDGSV + GGL RD+ G + AFV + F E+
Sbjct: 956 RSIIWCEWKKPEIGWTKLNTDGSVNKETAGFGGLLRDYRGEPICAFVSKAPQGDTFLVEL 1135
Query: 191 SGFILVMEHAALHGWYNIWLESDASSAL 218
+ + G +IW+ESD+ S +
Sbjct: 1136 WAIWRGLVLSLGLGIKSIWVESDSMSVV 1219
>BF643558
Length = 645
Score = 48.5 bits (114), Expect = 3e-06
Identities = 30/91 (32%), Positives = 41/91 (44%), Gaps = 3/91 (3%)
Frame = +2
Query: 76 RNNVRFSSAKVSSHAVQVRVHAFIGLSGAVSTGKCI---AADAAILDAFRIPPHRRSMRE 132
RN RF++ + + + + LSG T C D L F I H +
Sbjct: 365 RNQRRFNNKMIHWKSSITSIISSTFLSGNY-TSACDYTNITDFVFLKKFSIDIHPPKAPK 541
Query: 133 IVSVCWKPPSAPWVKVNTDGSVLNNSGACGG 163
I+ V W PP W+K NTDGS N+ +CGG
Sbjct: 542 IIEVLWNPPPLHWIKCNTDGSSNTNTSSCGG 634
>TC83780
Length = 400
Score = 33.5 bits (75), Expect(2) = 2e-04
Identities = 13/31 (41%), Positives = 17/31 (53%)
Frame = +3
Query: 1 MCSFCLEQVETSDHLFLRCKFVVTLWSWLCS 31
+C FC Q +S H+FL C + L WL S
Sbjct: 12 ICCFCHNQAVSSHHIFLSCPVTMVLCDWLSS 104
Score = 28.5 bits (62), Expect(2) = 2e-04
Identities = 14/36 (38%), Positives = 22/36 (60%)
Frame = +1
Query: 44 ALMSSLPRHCSSQVRDLYVAAVVHMVHSIWWARNNV 79
AL++S SS +R + + AV+H + IW RNN+
Sbjct: 136 ALITSCLSTSSSLLRKVMLFAVLHTIWIIWIERNNI 243
>BG586862
Length = 804
Score = 42.4 bits (98), Expect = 2e-04
Identities = 32/126 (25%), Positives = 53/126 (41%)
Frame = -1
Query: 1 MCSFCLEQVETSDHLFLRCKFVVTLWSWLCSQLHVGLDFSSIKALMSSLPRHCSSQVRDL 60
+C C ++ET HLFL C+ VT W SQL + S + + +
Sbjct: 543 LCPRCESKIETVQHLFLNCE--VTQKEWFGSQLGINFHSSGVLHFHDWITNFILKNDEET 370
Query: 61 YVAAVVHMVHSIWWARNNVRFSSAKVSSHAVQVRVHAFIGLSGAVSTGKCIAADAAILDA 120
+ A+ +++SIW ARN F + V V R S ++ + K ++L +
Sbjct: 369 -IIALTALLYSIWHARNQKVFENIDVPGDVVIQRA------SSSLHSFKMAQVSDSVLPS 211
Query: 121 FRIPPH 126
IP +
Sbjct: 210 NAIPSY 193
>TC78513 homologue to GP|5679340|emb|CAB51773.1 hypothetical protein {Galega
orientalis}, partial (20%)
Length = 765
Score = 41.2 bits (95), Expect = 5e-04
Identities = 23/78 (29%), Positives = 41/78 (52%), Gaps = 2/78 (2%)
Frame = +2
Query: 222 KNPSLVPILLRNRWHNARTL--NVQVISSHIFREGNVCVDRLANLGHSVVGEVWLSTLPS 279
K+ + +P L NRW+N + + ++ I SHI+REGN D LAN G S+ + + +P
Sbjct: 422 KSTNQIPWNL*NRWNNVKVILQGLKCIISHIYREGNQVADTLANHGLSLDSIHFWNDVPE 601
Query: 280 DFHQVFYEDRCSMPRIRY 297
++ ++ R+
Sbjct: 602 YTRASYFRNKDGWSNFRF 655
>TC83226 weakly similar to PIR|G86419|G86419 probable reverse transcriptase
100033-105622 [imported] - Arabidopsis thaliana, partial
(2%)
Length = 885
Score = 40.0 bits (92), Expect = 0.001
Identities = 52/234 (22%), Positives = 83/234 (35%), Gaps = 12/234 (5%)
Frame = +3
Query: 1 MCSFCLEQVETSDHLFLRCKFVVTLWSWLCSQLHVGLDFSSIKALMSSLPRHCSSQVRDL 60
+C C + ET HLF+ C +W S L + D +LP + + L
Sbjct: 240 LCPRCHSKTETITHLFMSCPLSKRVW--FGSNLCINFD---------NLPN--PNFIN*L 380
Query: 61 YVAA----------VVHMVHSIWWARNNVRFSSAKVSSHAVQVRVHAFIGLSGAVSTGKC 110
Y A + +++++W ARN + + R S +S K
Sbjct: 381 YEAIL*KDECITI*IAAIIYNLWHARNLSVLEDQTILEMDIIQRA------SNCISDYKQ 542
Query: 111 IAADAAILDAFRIPPHRRSMREIVSVCWKPPSAPWVKVNTDGSVLNN-SGACGGLFRDHL 169
A A R R + WK P+ VKVNTD ++ N+ G + RD +
Sbjct: 543 ANTQAPPSMARTGYDPRSQHRPAKNTKWKRPNLGLVKVNTDANLQNHGKWGLGIIIRDEV 722
Query: 170 GTFLGAFVGNL-GRCSVFDTEVSGFILVMEHAALHGWYNIWLESDASSALMVFK 222
G + A G + E + M A G+ + E D + + K
Sbjct: 723 GLVMAASTWETDGNDRALEAEAYALLTGMRFAKDCGFXKVXFEGDNEKLMKMVK 884
>TC82520
Length = 833
Score = 40.0 bits (92), Expect = 0.001
Identities = 32/101 (31%), Positives = 49/101 (47%), Gaps = 9/101 (8%)
Frame = +2
Query: 10 ETSDHLFLRCKFVVTLWSWLCSQLHVGL-------DFSSIKALMSSLPRHCSSQVRDLYV 62
ET+ HLFL C +LWS + LH+ L F M+ PR S ++ ++
Sbjct: 329 ETATHLFLHCDIFGSLWSHVLRWLHLLLVLPADIRQFFIQFTSMAGSPRFTHSFLQIMWF 508
Query: 63 AAVVHMVHSIWWARNNVRF--SSAKVSSHAVQVRVHAFIGL 101
A+ V +W RNN F S + S+ QV++H+F+ L
Sbjct: 509 AS----VWVLWKKRNNRVFQNSLSDPSTFVEQVKMHSFLWL 619
>AL382788 weakly similar to GP|11322386|emb glycine receptor betaZ subunit
{Danio rerio}, partial (4%)
Length = 353
Score = 39.7 bits (91), Expect = 0.001
Identities = 23/82 (28%), Positives = 31/82 (37%), Gaps = 2/82 (2%)
Frame = +2
Query: 142 SAPWVKVNTDGSVLNNS--GACGGLFRDHLGTFLGAFVGNLGRCSVFDTEVSGFILVMEH 199
S W+K D + +S G LFRD G+ G F+ G E +E
Sbjct: 2 SGGWIKAKIDNATCGSSDPSIVGSLFRDQFGSNFGCFIIFFGVNFAIHAEFYAAAYAVEI 181
Query: 200 AALHGWYNIWLESDASSALMVF 221
L W+N WL D + F
Sbjct: 182 VYLCNWHNFWLACDLKLVVDAF 247
>CB892607 similar to GP|8927657|gb| EST gb|N38213 comes from this gene.
{Arabidopsis thaliana}, partial (1%)
Length = 782
Score = 39.3 bits (90), Expect = 0.002
Identities = 22/55 (40%), Positives = 30/55 (54%), Gaps = 2/55 (3%)
Frame = +1
Query: 140 PPSAPWVKVNTDGSVLNN--SGACGGLFRDHLGTFLGAFVGNLGRCSVFDTEVSG 192
PP++ V + D VLN + ACGGL +D G F+ + LG CSV E+ G
Sbjct: 559 PPNSCEVALKCDYVVLNCDLNAACGGLIQDDQGHFVFHYANKLGSCSVLQAELWG 723
>TC93101
Length = 675
Score = 38.5 bits (88), Expect = 0.003
Identities = 24/91 (26%), Positives = 43/91 (46%)
Frame = -1
Query: 1 MCSFCLEQVETSDHLFLRCKFVVTLWSWLCSQLHVGLDFSSIKALMSSLPRHCSSQVRDL 60
+C C +ET++H+F+ C+ +W SQL + +S L S+Q ++
Sbjct: 615 LCPRCYFNLETTNHIFMSCERTQRVW--FGSQLSIRFPDNSTINFSDWLFDAISNQTEEI 442
Query: 61 YVAAVVHMVHSIWWARNNVRFSSAKVSSHAV 91
+ + + +SIW ARN F + VS +
Sbjct: 441 IIK-ISAITYSIWHARNKAIFENQFVSEDTI 352
>TC82948
Length = 705
Score = 38.1 bits (87), Expect = 0.004
Identities = 12/41 (29%), Positives = 22/41 (53%)
Frame = +3
Query: 1 MCSFCLEQVETSDHLFLRCKFVVTLWSWLCSQLHVGLDFSS 41
MCS C + E++ HLF C V +W W + +++ ++
Sbjct: 441 MCSLCFKSFESTHHLFFECSVSVQIWDWFSNLINLNFHINN 563
>TC83398 similar to PIR|T05150|T05150 hypothetical protein F18E5.40 -
Arabidopsis thaliana, partial (6%)
Length = 766
Score = 36.2 bits (82), Expect = 0.016
Identities = 30/113 (26%), Positives = 51/113 (44%), Gaps = 1/113 (0%)
Frame = +2
Query: 167 DHLGTFLGAFVGNLGRCS-VFDTEVSGFILVMEHAALHGWYNIWLESDASSALMVFKNPS 225
+H G F F G++ + + E+ ++ ++ A ++ SD+ + + PS
Sbjct: 80 EHFGFFNSGFSGHIDHSNDILFAELHAILMGLQLAQTLNIVDVVCYSDSLHYVNLINGPS 259
Query: 226 LVPILLRNRWHNARTLNVQVISSHIFREGNVCVDRLANLGHSVVGEVWLSTLP 278
+V + + L +++ H REGN C D LA LG SV STLP
Sbjct: 260 VVYHAYATLIQDIKDL-IRLSKLHTLREGNRCADFLAKLGASVD-----STLP 400
>BQ122106 similar to GP|9366656|emb|C probable similar to ring-h2 finger
protein rha1a. {Trypanosoma brucei}, partial (18%)
Length = 693
Score = 35.0 bits (79), Expect = 0.035
Identities = 46/148 (31%), Positives = 58/148 (39%), Gaps = 15/148 (10%)
Frame = -2
Query: 148 VNTDGSVLNNSGACG--GLFRDHLGTFLGAFVGNLGRCSVFDTEVSGFILVMEHAALHGW 205
+N DGS L N G GL R+ G F F GN+ T S +L HA G
Sbjct: 539 LNVDGSCLGNP*PTGFNGLIRNIAGLFNSGFPGNI-------TNTSDILLAELHAIFQGL 381
Query: 206 YNIWLESDASSALMVFKNPSLVPILLRN----RWHNARTLNVQ----VISS-----HIFR 252
I SD + V SL + L N ++H TL VI+S H
Sbjct: 380 RMI---SDMGISDFVCYFDSLHYVSLINGPSMKFHVYATLIQDIKDLVITSKASVFHTLC 210
Query: 253 EGNVCVDRLANLGHSVVGEVWLSTLPSD 280
EGN C D L LG + + + P D
Sbjct: 209 EGNYCADFLEMLGAASDSVLTIHVSPPD 126
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.326 0.137 0.449
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,926,741
Number of Sequences: 36976
Number of extensions: 190673
Number of successful extensions: 1341
Number of sequences better than 10.0: 60
Number of HSP's better than 10.0 without gapping: 1324
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1338
length of query: 298
length of database: 9,014,727
effective HSP length: 96
effective length of query: 202
effective length of database: 5,465,031
effective search space: 1103936262
effective search space used: 1103936262
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146708.4


