

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146708.3 - phase: 0
(282 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
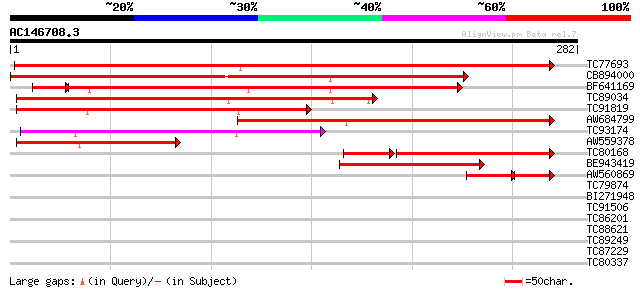
Score E
Sequences producing significant alignments: (bits) Value
TC77693 weakly similar to GP|7108597|gb|AAF36490.1| K+ transport... 407 e-114
CB894000 similar to GP|7108597|gb| K+ transporter HAK5 {Arabidop... 295 1e-80
BF641169 similar to GP|20196874|gb putative potassium transporte... 191 1e-52
TC89034 similar to PIR|T00487|T00487 probable potassium transpor... 184 4e-47
TC91819 similar to GP|17064932|gb|AAL32620.1 Similar to high aff... 155 1e-38
AW684799 similar to GP|17064932|gb Similar to high affinity pota... 139 9e-34
TC93174 weakly similar to GP|18129282|emb|CAD20319. putative pot... 124 4e-29
AW559378 similar to GP|9955527|emb potassium transport protein-l... 107 4e-24
TC80168 similar to PIR|T00487|T00487 probable potassium transpor... 87 1e-20
BE943419 similar to PIR|T51433|T514 probable cation transport pr... 65 2e-11
AW560869 similar to GP|7108597|gb|A K+ transporter HAK5 {Arabido... 47 1e-10
TC79874 similar to GP|17065608|gb|AAL33784.1 unknown protein {Ar... 31 0.47
BI271948 similar to GP|6498428|dbj Similar to Arabidopsis thalia... 31 0.61
TC91506 weakly similar to GP|15529153|gb|AAK97671.1 AT4g23490/F1... 30 0.80
TC86201 similar to PIR|T49003|T49003 protein kinase-like protein... 30 1.4
TC88621 similar to PIR|H84698|H84698 hypothetical protein At2g29... 29 1.8
TC89249 similar to GP|13486667|dbj|BAB39904. contains ESTs D4830... 29 2.3
TC87229 homologue to GP|16226927|gb|AAL16300.1 At1g63720/F24D7_9... 28 4.0
TC80337 similar to PIR|T47222|T47222 replication licensing facto... 27 8.8
>TC77693 weakly similar to GP|7108597|gb|AAF36490.1| K+ transporter HAK5
{Arabidopsis thaliana}, partial (67%)
Length = 2635
Score = 407 bits (1047), Expect = e-114
Identities = 199/273 (72%), Positives = 231/273 (83%), Gaps = 4/273 (1%)
Frame = +3
Query: 3 TLALAFQSLGVVYGDIGTSPLYVLASTFPKGIDHTDDILGVLSLIYYTILALPLLKYVFI 62
TL LAFQS+G+VYGDIGTSPLYV ASTF KGI + DDILGVLSLI YTI+ +P+LKYVFI
Sbjct: 216 TLTLAFQSIGIVYGDIGTSPLYVYASTFAKGIKNNDDILGVLSLIIYTIVLIPMLKYVFI 395
Query: 63 VLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSNQQ----LKKKL 118
VL ANDNGNGG FALYSL+CRH +SL PNQQPEDMELSNYKLE PSS Q+ LK+ L
Sbjct: 396 VLWANDNGNGGAFALYSLICRHIKMSLAPNQQPEDMELSNYKLEIPSSQQKRAHKLKQML 575
Query: 119 ENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISSKLGQDYVVSITIAILVIL 178
ENSHFAR++LL + I+GT MVIGDG+ TP +SV+SAV+GIS+ LGQ+ VV IT+AILV+L
Sbjct: 576 ENSHFARIILLLLAIMGTAMVIGDGILTPSISVLSAVSGISTSLGQNAVVGITVAILVVL 755
Query: 179 FCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYFQRNGKN 238
F QRFGT KVG FAP++ IWFI IG G+YN+FKYD+ VL A NPKYIVDYF+RNGK
Sbjct: 756 FSLQRFGTDKVGVLFAPVILIWFIFIGGIGLYNLFKYDIGVLKAFNPKYIVDYFKRNGKE 935
Query: 239 AWMSLGGVFLCISGCEAMFADLGHFNVRAIQVS 271
W+SLGGVFLCI+G EAMFADLGHF+VRAIQ+S
Sbjct: 936 GWLSLGGVFLCITGSEAMFADLGHFSVRAIQIS 1034
>CB894000 similar to GP|7108597|gb| K+ transporter HAK5 {Arabidopsis
thaliana}, partial (28%)
Length = 859
Score = 295 bits (755), Expect = 1e-80
Identities = 147/231 (63%), Positives = 183/231 (78%), Gaps = 3/231 (1%)
Frame = +1
Query: 1 MATLALAFQSLGVVYGDIGTSPLYVLASTFPKGIDHTDDILGVLSLIYYTILALPLLKYV 60
+ TL+LAFQSLG++YGDIGTSPLYV STFP GI + D+LG LSLI YTI + +KY+
Sbjct: 169 LGTLSLAFQSLGIIYGDIGTSPLYVYDSTFPDGISNKQDLLGCLSLIIYTISLIVFVKYI 348
Query: 61 FIVLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSNQQLKKKLEN 120
+VL ANDNGNGGT ALYSL+CRH+ VSLIPN QPED+E+S+YKLET S Q++K KLEN
Sbjct: 349 LVVLWANDNGNGGTCALYSLICRHSKVSLIPNHQPEDIEISHYKLET-RSRQKIKHKLEN 525
Query: 121 SHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGI---SSKLGQDYVVSITIAILVI 177
S FA++ L +TI+ T MVIGDG+ TP +SV+SAV+GI SS LGQ V+ I+I IL+I
Sbjct: 526 SKFAKLFLFIVTIMATAMVIGDGILTPSISVLSAVSGIRTRSSSLGQGAVLGISIGILII 705
Query: 178 LFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYI 228
LF AQRFGT KV ++FAPIL +WF+LIG G+YN+ K+D+ VL A NPKYI
Sbjct: 706 LFGAQRFGTDKVAYAFAPILLVWFLLIGGIGLYNLIKHDIGVLRAFNPKYI 858
>BF641169 similar to GP|20196874|gb putative potassium transporter
{Arabidopsis thaliana}, partial (26%)
Length = 681
Score = 191 bits (484), Expect(2) = 1e-52
Identities = 101/206 (49%), Positives = 135/206 (65%), Gaps = 10/206 (4%)
Frame = +1
Query: 30 FPKGIDHTD---DILGVLSLIYYTILALPLLKYVFIVLKANDNGNGGTFALYSLLCRHAN 86
F + I+H++ +I G LS +++T+ +PL KYVF+VL+A+DNG GGTFALYSL+CRHA
Sbjct: 58 FAEDIEHSETNEEIYGTLSFVFWTLTLIPLFKYVFVVLRADDNGEGGTFALYSLICRHAK 237
Query: 87 VSLIPNQQPEDMELSNYKLETPSSNQQLKKK--LENSHFARVLLLFMTILGTTMVIGDGV 144
VSL+PN+Q D LS+YK+E P + K K LE LL + +LGT MVIGDGV
Sbjct: 238 VSLLPNRQHADEALSSYKMEEPPEKEYSKFKMLLEKYKTLHTALLIVVLLGTCMVIGDGV 417
Query: 145 FTPPMSVISAVNGI-----SSKLGQDYVVSITIAILVILFCAQRFGTSKVGFSFAPILTI 199
TP +SV SAV+G+ S K Q V+ IT ILV LF Q +GT K GF FAPI+
Sbjct: 418 LTPAISVFSAVSGLEVSIMSKKHHQYAVIPITCFILVCLFALQHYGTHKGGFFFAPIVLT 597
Query: 200 WFILIGATGIYNVFKYDVRVLLAINP 225
W + I G+YN+FK++ V A++P
Sbjct: 598 WLLCIXTLGVYNIFKWNXHVYKALSP 675
Score = 33.5 bits (75), Expect(2) = 1e-52
Identities = 14/19 (73%), Positives = 15/19 (78%)
Frame = +3
Query: 12 GVVYGDIGTSPLYVLASTF 30
GVVYGD+ SPLYV STF
Sbjct: 3 GVVYGDLSISPLYVFTSTF 59
>TC89034 similar to PIR|T00487|T00487 probable potassium transport protein
F19I3.29 - Arabidopsis thaliana, partial (27%)
Length = 940
Score = 184 bits (466), Expect = 4e-47
Identities = 97/186 (52%), Positives = 128/186 (68%), Gaps = 6/186 (3%)
Frame = +2
Query: 4 LALAFQSLGVVYGDIGTSPLYVLASTFPKGIDHTDDILGVLSLIYYTILALPLLKYVFIV 63
L LA+QSLGVVYGD+GTSPLYV +TFP+G+D + ++G LSLI Y++ +PLLKYVFIV
Sbjct: 383 LRLAYQSLGVVYGDLGTSPLYVFYNTFPRGVDDPEKVIGALSLIIYSLTLVPLLKYVFIV 562
Query: 64 LKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLET---PSSNQQLKKKLEN 120
L+ANDNG GGT ALYSLLCRHAN+ +IPNQ D EL+ Y T S + ++ LE
Sbjct: 563 LRANDNGQGGTLALYSLLCRHANIKIIPNQHRTDEELTTYSRTTFHEKSFAAKTQRWLEE 742
Query: 121 SHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGIS-SKLGQDYVVSITIAILVI-- 177
F + +L + ++GT MVIGDG+ TP +SV+SA GI +K D V +A +++
Sbjct: 743 QKFIKSTILILVLVGTCMVIGDGILTPAISVLSAAGGIKVNKPEVDSGVXXLVAXVILXG 922
Query: 178 LFCAQR 183
LF QR
Sbjct: 923 LFXIQR 940
>TC91819 similar to GP|17064932|gb|AAL32620.1 Similar to high affinity
potassium transporter {Arabidopsis thaliana}, partial
(19%)
Length = 795
Score = 155 bits (393), Expect = 1e-38
Identities = 83/158 (52%), Positives = 108/158 (67%), Gaps = 11/158 (6%)
Frame = +1
Query: 4 LALAFQSLGVVYGDIGTSPLYVLASTFPKGIDHT---DDILGVLSLIYYTILALPLLKYV 60
L LA+QSLGVVYGD+ SPLYV STF +GI H+ ++I GVLSL+++++ +PL+KYV
Sbjct: 148 LTLAYQSLGVVYGDLSISPLYVFRSTFGEGIGHSNTNEEIYGVLSLVFWSVTLVPLVKYV 327
Query: 61 FIVLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSNQ-------- 112
FIVL+A+DNG GGTFALYSLLCR+A V+ +PN Q D ELS YK +
Sbjct: 328 FIVLRADDNGEGGTFALYSLLCRYAKVNSLPNCQLADEELSEYKKDGCGGGVSNGKGFAF 507
Query: 113 QLKKKLENSHFARVLLLFMTILGTTMVIGDGVFTPPMS 150
+LK LE + LL + ++GT MVIGDGV TP +S
Sbjct: 508 RLKSTLEKRKVLQKFLLVLALIGTCMVIGDGVLTPALS 621
>AW684799 similar to GP|17064932|gb Similar to high affinity potassium
transporter {Arabidopsis thaliana}, partial (21%)
Length = 585
Score = 139 bits (351), Expect = 9e-34
Identities = 71/162 (43%), Positives = 100/162 (60%), Gaps = 4/162 (2%)
Frame = +1
Query: 114 LKKKLENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISSKLGQDY----VVS 169
L LE + LL + ++GT MVIGDGV TP +SV SA++G + +++ V
Sbjct: 31 LSLHLEKRKVLQKFLLVLALIGTCMVIGDGVLTPALSVFSAISGFELSMSKEHHAYVEVP 210
Query: 170 ITIAILVILFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIV 229
+ ILV LF Q FGT +VGF FAPI+ W I A GIYN+F ++ ++ A+ P Y
Sbjct: 211 VACIILVGLFALQHFGTHRVGFMFAPIVMAWLFCISAIGIYNIFHWNSQIYRALCPIYAF 390
Query: 230 DYFQRNGKNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVS 271
+ ++N WM+LGGV L I+G EAMFADLGHF+ +IQ++
Sbjct: 391 RFMRQNQTGGWMALGGVLLXITGSEAMFADLGHFSQLSIQIA 516
>TC93174 weakly similar to GP|18129282|emb|CAD20319. putative potassium
transporter {Cymodocea nodosa}, partial (19%)
Length = 674
Score = 124 bits (311), Expect = 4e-29
Identities = 69/159 (43%), Positives = 94/159 (58%), Gaps = 7/159 (4%)
Frame = +1
Query: 6 LAFQSLGVVYGDIGTSPLYVLASTFP---KGIDHTDDILGVLSLIYYTILALPLLKYVFI 62
L +QSLG ++GDI SPLYV S F K + + D I G SLI++T+ + LLKY
Sbjct: 151 LTYQSLGFLFGDISLSPLYVYQSIFSGRLKHVQNEDAIFGSFSLIFWTLSLISLLKYAVF 330
Query: 63 VLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSN----QQLKKKL 118
+L A+DNG GGT ALYS LCR+A L+PN Q D ELS Y+ S+ LK+ +
Sbjct: 331 MLSADDNGEGGTVALYSHLCRNAKFCLLPNHQASDEELSTYRKPGYSNRNIPPSSLKRFI 510
Query: 119 ENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNG 157
E +++LL +LG M+I G P +SV+S+V G
Sbjct: 511 EKHKNTKIVLLIFVLLGACMIICVGALMPAISVLSSVEG 627
>AW559378 similar to GP|9955527|emb potassium transport protein-like
{Arabidopsis thaliana}, partial (15%)
Length = 610
Score = 107 bits (268), Expect = 4e-24
Identities = 51/83 (61%), Positives = 66/83 (79%), Gaps = 1/83 (1%)
Frame = +1
Query: 4 LALAFQSLGVVYGDIGTSPLYVLASTFPKG-IDHTDDILGVLSLIYYTILALPLLKYVFI 62
+ LAFQ+LGVV+GD+GTSPLY + F K I+ +DILG LSL+ YT++ +P LKYV +
Sbjct: 358 IVLAFQTLGVVFGDVGTSPLYTFSVMFRKAPINDNEDILGALSLVLYTLILIPFLKYVLV 537
Query: 63 VLKANDNGNGGTFALYSLLCRHA 85
VL AND+G GGTFALYSL+CR+A
Sbjct: 538 VLWANDDGEGGTFALYSLICRNA 606
>TC80168 similar to PIR|T00487|T00487 probable potassium transport protein
F19I3.29 - Arabidopsis thaliana, partial (48%)
Length = 1175
Score = 87.4 bits (215), Expect(2) = 1e-20
Identities = 37/79 (46%), Positives = 58/79 (72%)
Frame = +3
Query: 193 FAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYFQRNGKNAWMSLGGVFLCISG 252
FAPI+ +WF+LIG GIYN++++ VL A +P Y+ Y + K++W+SLGG+ L I+G
Sbjct: 87 FAPIVLLWFLLIGGIGIYNLWRFGGSVLRAFSPLYVYRYLRDGRKDSWLSLGGILLSITG 266
Query: 253 CEAMFADLGHFNVRAIQVS 271
EA+FADL +F V ++Q++
Sbjct: 267 TEALFADLANFPVSSVQIA 323
Score = 29.6 bits (65), Expect(2) = 1e-20
Identities = 13/25 (52%), Positives = 18/25 (72%)
Frame = +2
Query: 167 VVSITIAILVILFCAQRFGTSKVGF 191
VV + + ILV LF QR+GT +VG+
Sbjct: 8 VVLVAVVILVGLFSIQRYGTDRVGW 82
>BE943419 similar to PIR|T51433|T514 probable cation transport protein -
Arabidopsis thaliana, partial (9%)
Length = 449
Score = 65.5 bits (158), Expect = 2e-11
Identities = 29/72 (40%), Positives = 45/72 (62%)
Frame = +2
Query: 165 DYVVSITIAILVILFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAIN 224
DYVV ++ ILV LF Q GT +V F FAP++ W + I GIYN+F+++ +V A++
Sbjct: 230 DYVVIVSCIILVGLFSIQHHGTHRVAFMFAPVVAAWLLCISGIGIYNIFQWNRQVYRALS 409
Query: 225 PKYIVDYFQRNG 236
P Y+ + + G
Sbjct: 410 PVYMFRFLKTTG 445
>AW560869 similar to GP|7108597|gb|A K+ transporter HAK5 {Arabidopsis
thaliana}, partial (11%)
Length = 396
Score = 47.4 bits (111), Expect(2) = 1e-10
Identities = 19/25 (76%), Positives = 22/25 (88%)
Frame = +1
Query: 228 IVDYFQRNGKNAWMSLGGVFLCISG 252
IVDYF+RNGK W+SL GVFLCI+G
Sbjct: 1 IVDYFKRNGKEGWLSLSGVFLCITG 75
Score = 35.8 bits (81), Expect(2) = 1e-10
Identities = 16/20 (80%), Positives = 18/20 (90%)
Frame = +3
Query: 252 GCEAMFADLGHFNVRAIQVS 271
G EAMFADLGH +VRAIQ+S
Sbjct: 174 GSEAMFADLGHSSVRAIQIS 233
>TC79874 similar to GP|17065608|gb|AAL33784.1 unknown protein {Arabidopsis
thaliana}, partial (88%)
Length = 1154
Score = 31.2 bits (69), Expect = 0.47
Identities = 18/39 (46%), Positives = 25/39 (63%)
Frame = +3
Query: 54 LPLLKYVFIVLKANDNGNGGTFALYSLLCRHANVSLIPN 92
L L + +++ LKA+ GN GT AL +L R NVSL P+
Sbjct: 921 LALAQQLYVSLKAHGEGNLGTQALILVLERLNNVSLPPS 1037
>BI271948 similar to GP|6498428|dbj Similar to Arabidopsis thaliana
chromosome II BAC T27A16 sequence; hypothetical protein.
(AC005496), partial (33%)
Length = 695
Score = 30.8 bits (68), Expect = 0.61
Identities = 24/94 (25%), Positives = 42/94 (44%), Gaps = 8/94 (8%)
Frame = +3
Query: 124 ARVLLLFMTILGTTMVIGDGVFTPPMS--------VISAVNGISSKLGQDYVVSITIAIL 175
A++ L ++ ++ M IG+GV P M+ V ++ Y+ S+T +
Sbjct: 339 AKLGLPYLLVMRAFMGIGEGVAMPAMNNILSKWIPVSERSRSLALVYSGMYLGSVT-GLG 515
Query: 176 VILFCAQRFGTSKVGFSFAPILTIWFILIGATGI 209
F Q+FG V +SF + +IWF + I
Sbjct: 516 FSPFLIQKFGWPSVFYSFGSLGSIWFAFVXXKXI 617
>TC91506 weakly similar to GP|15529153|gb|AAK97671.1 AT4g23490/F16G20_190
{Arabidopsis thaliana}, partial (5%)
Length = 679
Score = 30.4 bits (67), Expect = 0.80
Identities = 14/25 (56%), Positives = 19/25 (76%), Gaps = 1/25 (4%)
Frame = -2
Query: 169 SITIAILVILF-CAQRFGTSKVGFS 192
S+TIA+ V+ F C+ R GT+ VGFS
Sbjct: 582 SVTIALSVVFFFCSHRLGTNAVGFS 508
>TC86201 similar to PIR|T49003|T49003 protein kinase-like protein -
Arabidopsis thaliana, partial (86%)
Length = 2127
Score = 29.6 bits (65), Expect = 1.4
Identities = 15/38 (39%), Positives = 20/38 (52%)
Frame = -3
Query: 215 YDVRVLLAINPKYIVDYFQRNGKNAWMSLGGVFLCISG 252
YD + A +PK V F + NA S+GG C+SG
Sbjct: 448 YDPSPINAFDPKLSVFSFSSSKDNAGTSIGGGAFCVSG 335
>TC88621 similar to PIR|H84698|H84698 hypothetical protein At2g29650
[imported] - Arabidopsis thaliana, partial (49%)
Length = 1074
Score = 29.3 bits (64), Expect = 1.8
Identities = 22/88 (25%), Positives = 39/88 (44%), Gaps = 8/88 (9%)
Frame = +1
Query: 124 ARVLLLFMTILGTTMVIGDGVFTPPMS--------VISAVNGISSKLGQDYVVSITIAIL 175
A++ L F+ + M IG+GV P M+ V ++ Y+ S+T +
Sbjct: 595 AKLGLPFLLVARAFMGIGEGVAMPAMNNILSKWVPVAERSRSLALVYSGMYLGSVT-GLA 771
Query: 176 VILFCAQRFGTSKVGFSFAPILTIWFIL 203
F ++G V +SF + T+WF +
Sbjct: 772 FSPFLIHQYGWPSVFYSFGSLGTVWFCI 855
>TC89249 similar to GP|13486667|dbj|BAB39904. contains ESTs D48306(S14443)
D24269(R1613) AU076096(E20048)~similar to Arabidopsis
thaliana, partial (78%)
Length = 1295
Score = 28.9 bits (63), Expect = 2.3
Identities = 31/137 (22%), Positives = 63/137 (45%), Gaps = 4/137 (2%)
Frame = +2
Query: 150 SVISAVNGISSK--LGQDYVVSITIAILVILFCAQRFGTSKVGFSFAPILTIWFILIGAT 207
S++ A G SK LG + +++A V + + S +GF FA LT W +++
Sbjct: 119 SILVAKMGEMSKTQLGVIGALFLSVASSVSIVICNKALMSNLGFPFATTLTSWHLMVTFC 298
Query: 208 GIYNVFKYDVRVLLAINPKYIVDYFQRNGKNAWMSLGGVFLCISGCEAMFADLGHFNVR- 266
++ ++++ ++ K ++ + NG +S+G + L + F +G + +
Sbjct: 299 TLHVALRFNLFEAKPVDMKTVMLFGILNG----VSIGFLNLSLG-----FNSIGFYQMTK 451
Query: 267 -AIQVSIIKLELTFVLK 282
AI + LE F+ K
Sbjct: 452 LAIIPFTVMLETLFLKK 502
>TC87229 homologue to GP|16226927|gb|AAL16300.1 At1g63720/F24D7_9
{Arabidopsis thaliana}, partial (27%)
Length = 1686
Score = 28.1 bits (61), Expect = 4.0
Identities = 42/165 (25%), Positives = 64/165 (38%), Gaps = 23/165 (13%)
Frame = +1
Query: 19 GTSPLYVLASTFPKGIDHTDDIL--GVLSLIYYTI-LALPL-LKYVFIVLKANDNGNGGT 74
GT P +++ S HT I +L+L ++ + L P Y+F+ L A D +
Sbjct: 37 GTQPFWLILS-------HTTQITRQSILNLRFHFLSLIFPC*AIYLFLFLYAADRSTSSS 195
Query: 75 FALYSLLCRHANVSLIPNQQPEDMELSNYKLET----------------PSSNQQLKKKL 118
F L + +H N+ + N+ D SN L+T PS+ KKK
Sbjct: 196 FFLIVKVRKHTNMRRVNNR--NDGRDSNNTLDTINAAAFAIASSHDRLQPSTATNQKKKW 369
Query: 119 ENSHFARVLLLFMTILGTTMVIGDGVFTP---PMSVISAVNGISS 160
N + + F IG V P P +A N +SS
Sbjct: 370 GN--WLNITGCFGYQKNNRKRIGHAVLVPETTPTGADAAANAVSS 498
>TC80337 similar to PIR|T47222|T47222 replication licensing factor MCM6
[validated] - African clawed frog, partial (26%)
Length = 1771
Score = 26.9 bits (58), Expect = 8.8
Identities = 18/56 (32%), Positives = 26/56 (46%)
Frame = +1
Query: 221 LAINPKYIVDYFQRNGKNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVSIIKLE 276
LA+ P Y+VD N + W S LC + +F LG F +Q ++ LE
Sbjct: 1408 LAVAPNYVVD**PSNDERVWKS-----LCFA--VPIFQRLGGFLCLELQACLVMLE 1554
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.326 0.143 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,663,825
Number of Sequences: 36976
Number of extensions: 122000
Number of successful extensions: 755
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 743
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 744
length of query: 282
length of database: 9,014,727
effective HSP length: 95
effective length of query: 187
effective length of database: 5,502,007
effective search space: 1028875309
effective search space used: 1028875309
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146708.3


