

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146708.15 - phase: 2
(392 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
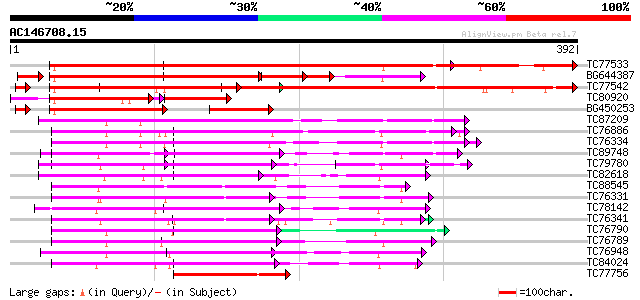
Score E
Sequences producing significant alignments: (bits) Value
TC77533 similar to GP|6996560|emb|CAB75429.1 oligouridylate bind... 535 e-153
BG644387 homologue to GP|6996560|emb oligouridylate binding prot... 266 2e-87
TC77542 similar to GP|6996560|emb|CAB75429.1 oligouridylate bind... 282 1e-76
TC80920 similar to GP|21553830|gb|AAM62923.1 oligouridylate bind... 148 8e-60
BG450253 similar to GP|21593280|gb oligouridylate binding protei... 120 9e-45
TC87209 similar to GP|9663767|emb|CAC01237.1 RNA Binding Protein... 128 4e-30
TC76886 similar to PIR|T01932|T01932 RNA binding protein homolog... 125 2e-29
TC76334 similar to GP|9663767|emb|CAC01237.1 RNA Binding Protein... 121 4e-28
TC89748 homologue to PIR|B84565|B84565 probable spliceosome asso... 99 2e-21
TC79780 similar to GP|18377731|gb|AAL67015.1 putative DNA bindin... 94 7e-20
TC82618 similar to GP|9294614|dbj|BAB02953.1 DNA/RNA binding pro... 92 3e-19
TC88545 similar to GP|13560783|gb|AAK30205.1 poly(A)-binding pro... 86 2e-17
TC76331 similar to GP|7673355|gb|AAF66823.1| poly(A)-binding pro... 85 5e-17
TC78142 similar to GP|7578881|gb|AAF64167.1| plastid-specific ri... 74 1e-13
TC76341 similar to GP|7528270|gb|AAF63202.1| poly(A)-binding pro... 73 2e-13
TC76790 similar to PIR|T06817|T06817 RNA-binding protein - garde... 70 1e-12
TC76789 similar to GP|15294254|gb|AAK95304.1 AT4g24770/F22K18_30... 70 2e-12
TC76948 similar to SP|Q08935|ROC1_NICSY 29 kDa ribonucleoprotein... 68 5e-12
TC84024 similar to PIR|S77714|S77714 RNA-binding protein precurs... 68 7e-12
TC77756 similar to PIR|T06796|T06796 glycine-rich RNA-binding pr... 67 2e-11
>TC77533 similar to GP|6996560|emb|CAB75429.1 oligouridylate binding protein
{Nicotiana plumbaginifolia}, partial (79%)
Length = 1621
Score = 535 bits (1379), Expect = e-153
Identities = 267/371 (71%), Positives = 303/371 (80%), Gaps = 6/371 (1%)
Frame = +1
Query: 28 CR--YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSSYGFIHYFDRRSAALAILTLN 85
CR YVGN+H QV+EPLLQE+F+ G +EGCKL RKEKSSYGF+ YFDR SAA+AI+TLN
Sbjct: 217 CRSVYVGNIHPQVSEPLLQELFSSAGALEGCKLIRKEKSSYGFVDYFDRSSAAIAIVTLN 396
Query: 86 GRHLFGQPIKVNWAYASGQREDTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMW 145
GR++FGQ IKVNWAY GQREDTSGH++IFVGDLSPEVTDATL+ACFS Y SCSDARVMW
Sbjct: 397 GRNIFGQSIKVNWAYTRGQREDTSGHFHIFVGDLSPEVTDATLYACFSAYSSCSDARVMW 576
Query: 146 DQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGGIEEKQNSDSKS 205
DQKTGRSRGFGFVSFR+QQ+AQSAINDLTGKWLGSRQIRCNWATKGA E Q+S+SKS
Sbjct: 577 DQKTGRSRGFGFVSFRNQQEAQSAINDLTGKWLGSRQIRCNWATKGANMNGENQSSESKS 756
Query: 206 VVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHALGAGVIEEVR 265
VVELT+G+SE+ +E++S+D PE NPQYTTVYVGNL E T +DLH HFHALG G IE+VR
Sbjct: 757 VVELTSGTSEEAQEMTSDDSPEKNPQYTTVYVGNLAPEVTSVDLHHHFHALGVGTIEDVR 936
Query: 266 VQRDKGFGFVRYSTHAEAALAIQMGNAQSYLCGKIIKCSWGSKPTPPGTASNPLPPPAA- 324
VQRDKGFGFVRYSTH EAALAIQMGN + +L GK IKCSWGSKPTPPGTAS PLPPPA+
Sbjct: 937 VQRDKGFGFVRYSTHGEAALAIQMGNTR-FLFGKPIKCSWGSKPTPPGTASTPLPPPAST 1113
Query: 325 -APLPGLSATDILAYERQLAMSKMGGVHAALMHPQAQHPLKQAA--IGASQAIYDGGFQN 381
P+PG S + YERQLA+SKM HA +K+AA +GA A Y GF N
Sbjct: 1114HVPVPGFSPAGLALYERQLALSKMNEAHA----------VKRAAMGMGALGAGYGAGFPN 1263
Query: 382 VAAAQQMMYYQ 392
VA Q +MYYQ
Sbjct: 1264VATTQHLMYYQ 1296
Score = 60.5 bits (145), Expect = 1e-09
Identities = 51/206 (24%), Positives = 88/206 (41%), Gaps = 4/206 (1%)
Frame = +1
Query: 107 DTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDA 166
D+S +++VG++ P+V++ L FS + +++ +K+ +GFV + + A
Sbjct: 205 DSSTCRSVYVGNIHPQVSEPLLQELFSSAGALEGCKLIRKEKS----SYGFVDYFDRSSA 372
Query: 167 QSAINDLTGKWLGSRQIRCNWATKGAGGIEEKQNSDSKSVVELTNGSSEDGKEISSNDVP 226
AI L G+ + + I+ NWA T G ED
Sbjct: 373 AIAIVTLNGRNIFGQSIKVNWA--------------------YTRGQRED---------- 462
Query: 227 ENNPQYTTVYVGNLGSEATQLDLHRHFHALG----AGVIEEVRVQRDKGFGFVRYSTHAE 282
+ ++VG+L E T L+ F A A V+ + + R +GFGFV + E
Sbjct: 463 --TSGHFHIFVGDLSPEVTDATLYACFSAYSSCSDARVMWDQKTGRSRGFGFVSFRNQQE 636
Query: 283 AALAIQMGNAQSYLCGKIIKCSWGSK 308
A AI +L + I+C+W +K
Sbjct: 637 AQSAIN-DLTGKWLGSRQIRCNWATK 711
Score = 32.0 bits (71), Expect = 0.42
Identities = 21/57 (36%), Positives = 28/57 (48%)
Frame = +2
Query: 6 VEICLLVLIQVLAAVSSLSVRKCRYVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKE 62
VEICLL LI+VLAAV + R+ P + F P++G LF K+
Sbjct: 182 VEICLLALIRVLAAVFMSATFILRF--------QSPCFKSCFQVLVPLKGASLFEKK 328
>BG644387 homologue to GP|6996560|emb oligouridylate binding protein
{Nicotiana plumbaginifolia}, partial (53%)
Length = 717
Score = 266 bits (679), Expect(4) = 2e-87
Identities = 126/150 (84%), Positives = 139/150 (92%), Gaps = 2/150 (1%)
Frame = +2
Query: 28 CR--YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSSYGFIHYFDRRSAALAILTLN 85
CR YVGN+H QVTEPLLQEVF+ TGP+EGCKL +KEKSSYGF+ YFDRRSAALAI+TLN
Sbjct: 116 CRSVYVGNIHPQVTEPLLQEVFSSTGPLEGCKLIKKEKSSYGFVDYFDRRSAALAIVTLN 295
Query: 86 GRHLFGQPIKVNWAYASGQREDTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMW 145
GR+LFGQPIKVNWAY S QREDTS H+NIFVGDLSPEVTDATL+ACFSVY SCSDA+VMW
Sbjct: 296 GRNLFGQPIKVNWAYTSAQREDTSSHFNIFVGDLSPEVTDATLYACFSVYPSCSDAKVMW 475
Query: 146 DQKTGRSRGFGFVSFRSQQDAQSAINDLTG 175
DQK+GRSRGFGFVSFR+QQ+AQSAINDL G
Sbjct: 476 DQKSGRSRGFGFVSFRNQQEAQSAINDLNG 565
Score = 61.6 bits (148), Expect(4) = 2e-87
Identities = 26/32 (81%), Positives = 31/32 (96%)
Frame = +3
Query: 175 GKWLGSRQIRCNWATKGAGGIEEKQNSDSKSV 206
GKWLGSRQIRCNWATKGAGGI++ ++SD+KSV
Sbjct: 564 GKWLGSRQIRCNWATKGAGGIDDIKSSDAKSV 659
Score = 53.5 bits (127), Expect = 1e-07
Identities = 47/185 (25%), Positives = 78/185 (41%), Gaps = 4/185 (2%)
Frame = +2
Query: 107 DTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDA 166
D+S +++VG++ P+VT+ L FS +++ +K+ +GFV + ++ A
Sbjct: 104 DSSTCRSVYVGNIHPQVTEPLLQEVFSSTGPLEGCKLIKKEKSS----YGFVDYFDRRSA 271
Query: 167 QSAINDLTGKWLGSRQIRCNWATKGAGGIEEKQNSDSKSVVELTNGSSEDGKEISSNDVP 226
AI L G+ L + I+ NWA A Q D+ S
Sbjct: 272 ALAIVTLNGRNLFGQPIKVNWAYTSA------QREDTSS--------------------- 370
Query: 227 ENNPQYTTVYVGNLGSEATQLDLHRHFHAL----GAGVIEEVRVQRDKGFGFVRYSTHAE 282
+ ++VG+L E T L+ F A V+ + + R +GFGFV + E
Sbjct: 371 -----HFNIFVGDLSPEVTDATLYACFSVYPSCSDAKVMWDQKSGRSRGFGFVSFRNQQE 535
Query: 283 AALAI 287
A AI
Sbjct: 536 AQSAI 550
Score = 34.7 bits (78), Expect = 0.065
Identities = 24/72 (33%), Positives = 37/72 (51%)
Frame = +2
Query: 234 TVYVGNLGSEATQLDLHRHFHALGAGVIEEVRVQRDKGFGFVRYSTHAEAALAIQMGNAQ 293
+VYVGN+ + T+ L F + G ++ + +GFV Y AALAI N +
Sbjct: 122 SVYVGNIHPQVTEPLLQEVFSSTGPLEGCKLIKKEKSSYGFVDYFDRRSAALAIVTLNGR 301
Query: 294 SYLCGKIIKCSW 305
+ L G+ IK +W
Sbjct: 302 N-LFGQPIKVNW 334
Score = 29.3 bits (64), Expect(4) = 2e-87
Identities = 15/18 (83%), Positives = 16/18 (88%)
Frame = +3
Query: 6 VEICLLVLIQVLAAVSSL 23
VEICLL LIQVLAAV +L
Sbjct: 81 VEICLLDLIQVLAAVCTL 134
Score = 26.6 bits (57), Expect(4) = 2e-87
Identities = 11/21 (52%), Positives = 17/21 (80%)
Frame = +1
Query: 204 KSVVELTNGSSEDGKEISSND 224
K +VELT+G+S+DG + +S D
Sbjct: 652 KVLVELTSGTSDDGHDKASED 714
>TC77542 similar to GP|6996560|emb|CAB75429.1 oligouridylate binding protein
{Nicotiana plumbaginifolia}, partial (74%)
Length = 1840
Score = 282 bits (722), Expect = 1e-76
Identities = 154/263 (58%), Positives = 189/263 (71%), Gaps = 17/263 (6%)
Frame = +1
Query: 147 QKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGGIE-EKQNSDSKS 205
Q R +G R QDAQSAIND+TGKWLG+RQIRCNWATKGAGG E++ +DS++
Sbjct: 739 QNWSLKRIWGLFLSRDHQDAQSAINDMTGKWLGNRQIRCNWATKGAGGSSNEEKINDSQN 918
Query: 206 VVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHALGAGVIEEVR 265
V LTNGSS+ G++ S+ D PENNP YTTVYVGNL + TQ +LH FHALGAGV+EEVR
Sbjct: 919 AVVLTNGSSDGGQDNSNEDAPENNPSYTTVYVGNLPHDVTQAELHCQFHALGAGVLEEVR 1098
Query: 266 VQRDKGFGFVRYSTHAEAALAIQMGNAQSYLCGKIIKCSWGSKPTPPGTASNPLPPPAAA 325
VQ KGFGFVRY+TH EAA+AIQM N + + GK +KCSWGSKPTPPGTASNPLPPPAA
Sbjct: 1099VQSGKGFGFVRYNTHEEAAMAIQMANGRP-VRGKTMKCSWGSKPTPPGTASNPLPPPAAQ 1275
Query: 326 P---LP------GLSATDILAYERQLAMSK---MGGVHAALMHPQAQHPLKQAAIG---- 369
P LP G +A ++LAY+RQLA+S+ G AL+ QH L A++G
Sbjct: 1276PYQILPTAGMNQGYTAAELLAYQRQLALSQAAVSGLSGQALLQMSGQHGLAPASMGINSA 1455
Query: 370 ASQAIYDGGFQNVAAAQQMMYYQ 392
ASQA+YD G+ ++ QQ+MYY+
Sbjct: 1456ASQAMYD-GYAGNSSRQQLMYYR 1521
Score = 210 bits (535), Expect(2) = 8e-55
Identities = 98/135 (72%), Positives = 112/135 (82%), Gaps = 2/135 (1%)
Frame = +2
Query: 28 CR--YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSSYGFIHYFDRRSAALAILTLN 85
CR YVGN+H VT+ LL EVF GP+ GCKL RKEKSSYGF+ Y DR SAALAI+TL+
Sbjct: 374 CRSVYVGNIHVNVTDKLLAEVFQSAGPLAGCKLIRKEKSSYGFVDYHDRASAALAIMTLH 553
Query: 86 GRHLFGQPIKVNWAYASGQREDTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMW 145
GR L+GQ +KVNWAYA+ REDTSGH+N+FVGDLSPEVTDATLFACFSVY +CSDARVMW
Sbjct: 554 GRQLYGQALKVNWAYANSSREDTSGHFNVFVGDLSPEVTDATLFACFSVYTTCSDARVMW 733
Query: 146 DQKTGRSRGFGFVSF 160
D KTGRS+G+G F
Sbjct: 734 DHKTGRSKGYGVCFF 778
Score = 45.8 bits (107), Expect = 3e-05
Identities = 37/161 (22%), Positives = 64/161 (38%), Gaps = 33/161 (20%)
Frame = +1
Query: 63 KSSYGFIHYFDRRSAALAILTLNGRHLFGQPIKVNWAY--ASGQR--------------- 105
K +G D + A AI + G+ L + I+ NWA A G
Sbjct: 754 KRIWGLFLSRDHQDAQSAINDMTGKWLGNRQIRCNWATKGAGGSSNEEKINDSQNAVVLT 933
Query: 106 ----------------EDTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKT 149
E+ + ++VG+L +VT A L F A V+ + +
Sbjct: 934 NGSSDGGQDNSNEDAPENNPSYTTVYVGNLPHDVTQAELHCQFHAL----GAGVLEEVRV 1101
Query: 150 GRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATK 190
+GFGFV + + ++A AI G+ + + ++C+W +K
Sbjct: 1102QSGKGFGFVRYNTHEEAAMAIQMANGRPVRGKTMKCSWGSK 1224
Score = 40.4 bits (93), Expect = 0.001
Identities = 40/171 (23%), Positives = 65/171 (37%), Gaps = 4/171 (2%)
Frame = +2
Query: 107 DTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDA 166
D S +++VG++ VTD L F + +++ +K+ +GFV + + A
Sbjct: 362 DASACRSVYVGNIHVNVTDKLLAEVFQSAGPLAGCKLIRKEKSS----YGFVDYHDRASA 529
Query: 167 QSAINDLTGKWLGSRQIRCNWATKGAGGIEEKQNSDSKSVVELTNGSSEDGKEISSNDVP 226
AI L G+ L + ++ NWA N S ED
Sbjct: 530 ALAIMTLHGRQLYGQALKVNWA--------------------YANSSRED---------- 619
Query: 227 ENNPQYTTVYVGNLGSEATQLDLHRHFHAL----GAGVIEEVRVQRDKGFG 273
+ V+VG+L E T L F A V+ + + R KG+G
Sbjct: 620 --TSGHFNVFVGDLSPEVTDATLFACFSVYTTCSDARVMWDHKTGRSKGYG 766
Score = 39.3 bits (90), Expect = 0.003
Identities = 24/71 (33%), Positives = 37/71 (51%), Gaps = 2/71 (2%)
Frame = +1
Query: 30 YVGNVHTQVTEPLLQEVF--AGTGPVEGCKLFRKEKSSYGFIHYFDRRSAALAILTLNGR 87
YVGN+ VT+ L F G G +E ++ + +GF+ Y AA+AI NGR
Sbjct: 1009 YVGNLPHDVTQAELHCQFHALGAGVLEEVRV--QSGKGFGFVRYNTHEEAAMAIQMANGR 1182
Query: 88 HLFGQPIKVNW 98
+ G+ +K +W
Sbjct: 1183 PVRGKTMKCSW 1215
Score = 33.9 bits (76), Expect = 0.11
Identities = 23/72 (31%), Positives = 35/72 (47%)
Frame = +2
Query: 234 TVYVGNLGSEATQLDLHRHFHALGAGVIEEVRVQRDKGFGFVRYSTHAEAALAIQMGNAQ 293
+VYVGN+ T L F + G ++ + +GFV Y A AALAI + +
Sbjct: 380 SVYVGNIHVNVTDKLLAEVFQSAGPLAGCKLIRKEKSSYGFVDYHDRASAALAIMTLHGR 559
Query: 294 SYLCGKIIKCSW 305
L G+ +K +W
Sbjct: 560 Q-LYGQALKVNW 592
Score = 21.6 bits (44), Expect(2) = 8e-55
Identities = 9/10 (90%), Positives = 10/10 (100%)
Frame = +3
Query: 5 QVEICLLVLI 14
QVEICLLVL+
Sbjct: 336 QVEICLLVLM 365
>TC80920 similar to GP|21553830|gb|AAM62923.1 oligouridylate binding protein
putative {Arabidopsis thaliana}, partial (41%)
Length = 694
Score = 148 bits (374), Expect(2) = 8e-60
Identities = 72/74 (97%), Positives = 72/74 (97%), Gaps = 2/74 (2%)
Frame = +3
Query: 28 CR--YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSSYGFIHYFDRRSAALAILTLN 85
CR YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSSYGFIHYFDRRSAALAILTLN
Sbjct: 309 CRSVYVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSSYGFIHYFDRRSAALAILTLN 488
Query: 86 GRHLFGQPIKVNWA 99
GRHLFGQPIKVNWA
Sbjct: 489 GRHLFGQPIKVNWA 530
Score = 100 bits (248), Expect(2) = 8e-60
Identities = 46/46 (100%), Positives = 46/46 (100%)
Frame = +2
Query: 108 TSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSR 153
TSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSR
Sbjct: 557 TSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSR 694
Score = 45.1 bits (105), Expect = 5e-05
Identities = 42/121 (34%), Positives = 56/121 (45%), Gaps = 14/121 (11%)
Frame = +1
Query: 1 LNQSQVEICLLVLIQVLAAVSSLSVRKCRYVGNVHTQVTEPLLQEVFAGTGPVEGCKLFR 60
LNQSQVEICLLVLIQVLAAV C + ++H P CK F
Sbjct: 259 LNQSQVEICLLVLIQVLAAV-------CMWATSIH--------------K*PSHFCKKFL 375
Query: 61 KEKSSYGFIHYFDRRS----------AALAI----LTLNGRHLFGQPIKVNWAYASGQRE 106
+ + +++ +RS A L + L++ G + G K+ YASGQRE
Sbjct: 376 RVLAQLKGVNFLGKRSHPMDSFITSIADLLLWPY*LSMEGIY-SGNLSKLTGHYASGQRE 552
Query: 107 D 107
D
Sbjct: 553 D 555
Score = 38.1 bits (87), Expect = 0.006
Identities = 26/74 (35%), Positives = 41/74 (55%), Gaps = 2/74 (2%)
Frame = +3
Query: 234 TVYVGNLGSEATQLDLHRHFHALGAGVIEEVRVQRDK--GFGFVRYSTHAEAALAIQMGN 291
+VYVGN+ ++ T+ L F G G +E ++ R + +GF+ Y AALAI N
Sbjct: 315 SVYVGNVHTQVTEPLLQEVF--AGTGPVEGCKLFRKEKSSYGFIHYFDRRSAALAILTLN 488
Query: 292 AQSYLCGKIIKCSW 305
+ +L G+ IK +W
Sbjct: 489 GR-HLFGQPIKVNW 527
Score = 35.0 bits (79), Expect = 0.050
Identities = 21/82 (25%), Positives = 41/82 (49%)
Frame = +3
Query: 107 DTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDA 166
D S +++VG++ +VT+ L F+ ++ +K+ +GF+ + ++ A
Sbjct: 297 DPSTCRSVYVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSS----YGFIHYFDRRSA 464
Query: 167 QSAINDLTGKWLGSRQIRCNWA 188
AI L G+ L + I+ NWA
Sbjct: 465 ALAILTLNGRHLFGQPIKVNWA 530
>BG450253 similar to GP|21593280|gb oligouridylate binding protein putative
{Arabidopsis thaliana}, partial (40%)
Length = 680
Score = 120 bits (301), Expect(3) = 9e-45
Identities = 57/84 (67%), Positives = 66/84 (77%), Gaps = 2/84 (2%)
Frame = +3
Query: 28 CR--YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSSYGFIHYFDRRSAALAILTLN 85
CR YVGN+H VT+ LL EVF GP+ GCKL RKEKSSYGF+ Y DR SAALAI+TL+
Sbjct: 297 CRSVYVGNIHVNVTDKLLAEVFQSAGPLAGCKLIRKEKSSYGFVDYHDRASAALAIMTLH 476
Query: 86 GRHLFGQPIKVNWAYASGQREDTS 109
GR L+GQ +KVNWAYA+ REDTS
Sbjct: 477 GRQLYGQALKVNWAYANSSREDTS 548
Score = 77.0 bits (188), Expect(3) = 9e-45
Identities = 34/44 (77%), Positives = 38/44 (86%)
Frame = +2
Query: 139 SDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQ 182
SDARVMWD KTGRS+ +GFVSFR DAQSA ND+TGKWLG+RQ
Sbjct: 548 SDARVMWDHKTGRSKXYGFVSFRDHXDAQSAXNDMTGKWLGNRQ 679
Score = 38.1 bits (87), Expect = 0.006
Identities = 22/82 (26%), Positives = 40/82 (47%)
Frame = +3
Query: 107 DTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDA 166
D S +++VG++ VTD L F + +++ +K+ +GFV + + A
Sbjct: 285 DASACRSVYVGNIHVNVTDKLLAEVFQSAGPLAGCKLIRKEKSS----YGFVDYHDRASA 452
Query: 167 QSAINDLTGKWLGSRQIRCNWA 188
AI L G+ L + ++ NWA
Sbjct: 453 ALAIMTLHGRQLYGQALKVNWA 518
Score = 33.9 bits (76), Expect = 0.11
Identities = 23/72 (31%), Positives = 35/72 (47%)
Frame = +3
Query: 234 TVYVGNLGSEATQLDLHRHFHALGAGVIEEVRVQRDKGFGFVRYSTHAEAALAIQMGNAQ 293
+VYVGN+ T L F + G ++ + +GFV Y A AALAI + +
Sbjct: 303 SVYVGNIHVNVTDKLLAEVFQSAGPLAGCKLIRKEKSSYGFVDYHDRASAALAIMTLHGR 482
Query: 294 SYLCGKIIKCSW 305
L G+ +K +W
Sbjct: 483 Q-LYGQALKVNW 515
Score = 21.6 bits (44), Expect(3) = 9e-45
Identities = 9/10 (90%), Positives = 10/10 (100%)
Frame = +1
Query: 5 QVEICLLVLI 14
QVEICLLVL+
Sbjct: 259 QVEICLLVLM 288
>TC87209 similar to GP|9663767|emb|CAC01237.1 RNA Binding Protein 45
{Nicotiana plumbaginifolia}, partial (69%)
Length = 1504
Score = 128 bits (321), Expect = 4e-30
Identities = 92/307 (29%), Positives = 154/307 (49%), Gaps = 9/307 (2%)
Frame = +3
Query: 21 SSLSVRKCRYVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSS----YGFIHYFDRRS 76
SS K ++G++ + E L F+ TG V K+ R ++++ YGF+ + R
Sbjct: 240 SSADEVKTLWIGDLQYWMDENYLYNCFSHTGEVGSVKVIRNKQTNQSEGYGFLEFISRAG 419
Query: 77 AALAILTLNGRHL--FGQPIKVNWA-YASGQ-REDTSGHYNIFVGDLSPEVTDATLFACF 132
A + T NG + GQ ++NWA ++SG+ R D S Y IFVGDL+ +V+D L F
Sbjct: 420 AERVLQTFNGTIMPNGGQNFRLNWATFSSGEKRHDDSPDYTIFVGDLAADVSDHHLTEVF 599
Query: 133 SV-YQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKG 191
Y S A+V+ D+ TGR++G+GFV F + + A+ ++ G +R +R A+
Sbjct: 600 RTRYNSVKGAKVVIDRTTGRTKGYGFVRFADESEQMRAMTEMQGVLCSTRPMRIGPASNK 779
Query: 192 AGGIEEKQNSDSKSVVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHR 251
G + SK+ + G +++ EN+P TT++VGNL T L +
Sbjct: 780 NLGTQ-----TSKASYQNPQGGAQN----------ENDPNNTTIFVGNLDPNVTDEHLKQ 914
Query: 252 HFHALGAGVIEEVRVQRDKGFGFVRYSTHAEAALAIQMGNAQSYLCGKIIKCSWGSKPTP 311
F G + V++ K GFV+++ + A A+++ N + L G+ ++ SWG P
Sbjct: 915 VFTQYGE--LVHVKIPSGKRCGFVQFADRSSAEEALRVLNG-TLLGGQNVRLSWGRSPAN 1085
Query: 312 PGTASNP 318
T +P
Sbjct: 1086KQTQQDP 1106
>TC76886 similar to PIR|T01932|T01932 RNA binding protein homolog - common
tobacco (fragment), partial (67%)
Length = 1896
Score = 125 bits (315), Expect = 2e-29
Identities = 90/295 (30%), Positives = 147/295 (49%), Gaps = 15/295 (5%)
Frame = +2
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSS----YGFIHYFDRRSAALAILTLN 85
++G++H + E L FA TG V K+ R +++ YGF+ ++ R A + N
Sbjct: 557 WLGDLHHWMDETFLHNCFAHTGEVASAKVIRNKQTGQSEGYGFVEFYTRAMAEKVLQNFN 736
Query: 86 GRHL--FGQPIKVNWAYAS-----GQRE--DTSGHYNIFVGDLSPEVTDATLFACF-SVY 135
G + Q ++NWA S G+R + + ++FVGDL+ +VTDA L F S +
Sbjct: 737 GTMMPNTDQAFRLNWATFSAAGGGGERRSSEATSDLSVFVGDLAIDVTDAMLQETFASKF 916
Query: 136 QSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWAT-KGAGG 194
S A+V+ D TGRS+G+GFV F + + A+ ++ G + SR +R AT K G
Sbjct: 917 SSIKGAKVVIDSNTGRSKGYGFVRFGDESERTRAMTEMNGVYCSSRPMRVGVATPKKTYG 1096
Query: 195 IEEKQNSDSKSVVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFH 254
++ +S + V L G ++ E + TT++VG L S+ + DL + F
Sbjct: 1097NPQQYSSQA---VVLAGGGGHSNGAMAQGSQSEGDSNNTTIFVGGLDSDISDEDLRQPF- 1264
Query: 255 ALGAGVIEEVRVQRDKGFGFVRYSTHAEAALAIQMGNAQSYLCGKIIKCSWGSKP 309
L G + V++ KG GFV+ + A AIQ G + + + ++ SWG P
Sbjct: 1265-LQFGDVISVKIPVGKGCGFVQLADRKNAEEAIQ-GLNGTVIGKQTVRLSWGRSP 1423
Score = 68.2 bits (165), Expect = 5e-12
Identities = 59/219 (26%), Positives = 99/219 (44%), Gaps = 14/219 (6%)
Frame = +2
Query: 114 IFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDL 173
I++GDL + + L CF+ + A+V+ +++TG+S G+GFV F ++ A+ + +
Sbjct: 554 IWLGDLHHWMDETFLHNCFAHTGEVASAKVIRNKQTGQSEGYGFVEFYTRAMAEKVLQNF 733
Query: 174 TGKWLGS--RQIRCNWAT-KGAGGIEEKQNSDSKSVVELTNGSSEDGKEISSNDVPENNP 230
G + + + R NWAT AGG E+++S++ S +
Sbjct: 734 NGTMMPNTDQAFRLNWATFSAAGGGGERRSSEATSDL----------------------- 844
Query: 231 QYTTVYVGNLGSEATQLDLHRHFHA-----LGAGVIEEVRVQRDKGFGFVRYSTHAEAAL 285
+V+VG+L + T L F + GA V+ + R KG+GFVR+ +E
Sbjct: 845 ---SVFVGDLAIDVTDAMLQETFASKFSSIKGAKVVIDSNTGRSKGYGFVRFGDESERTR 1015
Query: 286 AIQMGNAQSYLCGKIIKCSWGSKP------TPPGTASNP 318
A+ N + CS S+P TP T NP
Sbjct: 1016AMTEMNG--------VYCS--SRPMRVGVATPKKTYGNP 1102
Score = 37.4 bits (85), Expect = 0.010
Identities = 23/85 (27%), Positives = 44/85 (51%), Gaps = 3/85 (3%)
Frame = +2
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSSYGFIHYFDRRSAALAILTLNGRHL 89
+VG + + +++ L++ F G V K+ GF+ DR++A AI LNG +
Sbjct: 1211 FVGGLDSDISDEDLRQPFLQFGDVISVKI--PVGKGCGFVQLADRKNAEEAIQGLNGTVI 1384
Query: 90 FGQPIKVNWAYASGQ---REDTSGH 111
Q ++++W + G R D++G+
Sbjct: 1385 GKQTVRLSWGRSPGNKHWRNDSNGY 1459
>TC76334 similar to GP|9663767|emb|CAC01237.1 RNA Binding Protein 45
{Nicotiana plumbaginifolia}, partial (68%)
Length = 1579
Score = 121 bits (304), Expect = 4e-28
Identities = 91/300 (30%), Positives = 148/300 (49%), Gaps = 11/300 (3%)
Frame = +1
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSS----YGFIHYFDRRSAALAILTLN 85
++G++ + E L F TG + K+ R +++S YGFI + R +A + T N
Sbjct: 238 WIGDLQYWMDENYLYTCFGNTGELTSVKVIRNKQTSQSEGYGFIEFNTRATAERVLQTYN 417
Query: 86 GRHL--FGQPIKVNWA-YASGQR---EDTSGHYNIFVGDLSPEVTDATLFACFSV-YQSC 138
G + GQ ++NWA +++G+R +D + IFVGDL+ +VTD L F Y S
Sbjct: 418 GTIMPNGGQNYRLNWATFSAGERSSRQDDGPDHTIFVGDLAADVTDYLLQETFRARYNSV 597
Query: 139 SDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGGIEEK 198
A+V+ D+ TGRS+G+GFV F + + A+ ++ G +R +R AT
Sbjct: 598 KGAKVVIDRLTGRSKGYGFVRFADEGEQMRAMTEMQGVLCSTRPMRIGPATNK----NPA 765
Query: 199 QNSDSKSVVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHALGA 258
+ K+ + G S EN+P TT++VGNL T L + F G
Sbjct: 766 ATTQPKASYNPSGGQS------------ENDPNNTTIFVGNLDPNVTDDHLRQVFTQYGE 909
Query: 259 GVIEEVRVQRDKGFGFVRYSTHAEAALAIQMGNAQSYLCGKIIKCSWGSKPTPPGTASNP 318
+ V++ K GFV++S + A AI++ N + L G+ ++ SWG P+ T +P
Sbjct: 910 --LVHVKIPSGKRCGFVQFSDRSCAEEAIRVLNG-TLLGGQNVRLSWGRTPSNKQTQQDP 1080
Score = 71.2 bits (173), Expect = 6e-13
Identities = 56/220 (25%), Positives = 94/220 (42%), Gaps = 7/220 (3%)
Frame = +1
Query: 114 IFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDL 173
+++GDL + + L+ CF + +V+ +++T +S G+GF+ F ++ A+ +
Sbjct: 235 LWIGDLQYWMDENYLYTCFGNTGELTSVKVIRNKQTSQSEGYGFIEFNTRATAERVLQTY 414
Query: 174 TGKWL--GSRQIRCNWATKGAGGIEEKQNSDSKSVVELTNGSSEDGKEISSNDVPENNPQ 231
G + G + R NWAT AG +Q +D P++
Sbjct: 415 NGTIMPNGGQNYRLNWATFSAGERSSRQ-----------------------DDGPDH--- 516
Query: 232 YTTVYVGNLGSEATQLDLHRHFHA-----LGAGVIEEVRVQRDKGFGFVRYSTHAEAALA 286
T++VG+L ++ T L F A GA V+ + R KG+GFVR++ E A
Sbjct: 517 --TIFVGDLAADVTDYLLQETFRARYNSVKGAKVVIDRLTGRSKGYGFVRFADEGEQMRA 690
Query: 287 IQMGNAQSYLCGKIIKCSWGSKPTPPGTASNPLPPPAAAP 326
M Q LC ++P G A+N P P
Sbjct: 691 --MTEMQGVLC--------STRPMRIGPATNKNPAATTQP 780
>TC89748 homologue to PIR|B84565|B84565 probable spliceosome associated
protein [imported] - Arabidopsis thaliana, partial (56%)
Length = 707
Score = 99.4 bits (246), Expect = 2e-21
Identities = 59/166 (35%), Positives = 94/166 (56%), Gaps = 5/166 (3%)
Frame = +2
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFR----KEKSSYGFIHYFDRRSAALAILTLN 85
YVGN+ Q++E LL E+F GPV + + + YGF+ + A AI LN
Sbjct: 173 YVGNLDPQISEELLWELFVQAGPVVNVYVPKDRVTNQHQGYGFVEFRSEEDADYAIKVLN 352
Query: 86 GRHLFGQPIKVNWAYASGQREDTSGHYNIFVGDLSPEVTDATLFACFSVY-QSCSDARVM 144
L+G+PI+VN AS ++ N+F+G+L P+V + L+ FS + ++ ++M
Sbjct: 353 MIKLYGKPIRVN--KASQDKKSLDVGANLFIGNLDPDVDEKLLYDTFSAFGVIVTNPKIM 526
Query: 145 WDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATK 190
D T SRGFGF+S+ S + + SAI + G++L +RQI ++A K
Sbjct: 527 RDPDTRNSRGFGFISYDSFEASDSAIEAMNGQYLCNRQITVSYAYK 664
Score = 67.8 bits (164), Expect = 7e-12
Identities = 57/205 (27%), Positives = 91/205 (43%), Gaps = 6/205 (2%)
Frame = +2
Query: 115 FVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLT 174
+VG+L P++++ L+ F + V D+ T + +G+GFV FRS++DA AI L
Sbjct: 173 YVGNLDPQISEELLWELFVQAGPVVNVYVPKDRVTNQHQGYGFVEFRSEEDADYAIKVLN 352
Query: 175 GKWLGSRQIRCNWATKGAGGIEEKQNSDSKSVVELTNGSSEDGKEISSNDVPENNPQYTT 234
L + IR N K + D KS+ DV N
Sbjct: 353 MIKLYGKPIRVN-----------KASQDKKSL-----------------DVGAN------ 430
Query: 235 VYVGNLGSEATQLDLHRHFHALGAGVIEEVRVQRD------KGFGFVRYSTHAEAALAIQ 288
+++GNL + + L+ F A G ++ ++ RD +GFGF+ Y + + AI+
Sbjct: 431 LFIGNLDPDVDEKLLYDTFSAFGV-IVTNPKIMRDPDTRNSRGFGFISYDSFEASDSAIE 607
Query: 289 MGNAQSYLCGKIIKCSWGSKPTPPG 313
N Q YLC + I S+ K G
Sbjct: 608 AMNGQ-YLCNRQITVSYAYKKDTKG 679
Score = 42.7 bits (99), Expect = 2e-04
Identities = 30/94 (31%), Positives = 48/94 (50%), Gaps = 5/94 (5%)
Frame = +2
Query: 22 SLSVRKCRYVGNVHTQVTEPLLQEVFAGTGP-VEGCKLFR----KEKSSYGFIHYFDRRS 76
SL V ++GN+ V E LL + F+ G V K+ R + +GFI Y +
Sbjct: 410 SLDVGANLFIGNLDPDVDEKLLYDTFSAFGVIVTNPKIMRDPDTRNSRGFGFISYDSFEA 589
Query: 77 AALAILTLNGRHLFGQPIKVNWAYASGQREDTSG 110
+ AI +NG++L + I V++AY ++DT G
Sbjct: 590 SDSAIEAMNGQYLCNRQITVSYAY----KKDTKG 679
>TC79780 similar to GP|18377731|gb|AAL67015.1 putative DNA binding protein
ACBF {Arabidopsis thaliana}, partial (67%)
Length = 1649
Score = 94.4 bits (233), Expect = 7e-20
Identities = 58/173 (33%), Positives = 93/173 (53%), Gaps = 9/173 (5%)
Frame = +3
Query: 21 SSLSVRKCRYVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRK----EKSSYGFIHYFDRRS 76
+SL + ++G++ V E L F+ TG V K+ R + YGFI + +
Sbjct: 81 ASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSA 260
Query: 77 AALAILTLNGRHLFG--QPIKVNWA-YASGQREDTSG-HYNIFVGDLSPEVTDATLFACF 132
A + T NG + G Q ++NWA + G+R +G ++IFVGDL+P+VTD L F
Sbjct: 261 AERVLQTYNGTQMPGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDLAPDVTDYLLQETF 440
Query: 133 SV-YQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIR 184
Y S A+V+ D TGRS+G+GFV F + + A++++ G + +R +R
Sbjct: 441 RTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMR 599
Score = 56.6 bits (135), Expect = 2e-08
Identities = 47/185 (25%), Positives = 80/185 (42%), Gaps = 7/185 (3%)
Frame = +3
Query: 114 IFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDL 173
+++GDL V + L CFS +++ ++ TG+ G+GF+ F S A+ +
Sbjct: 105 LWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQTY 284
Query: 174 TGKWL-GSRQ-IRCNWATKGAGGIEEKQNSDSKSVVELTNGSSEDGKEISSNDVPENNPQ 231
G + G+ Q R NWA+ G G E++ P+ P
Sbjct: 285 NGTQMPGTEQTFRLNWASFGIG---ERR--------------------------PDAGPD 377
Query: 232 YTTVYVGNLGSEATQLDLHRHFHA-----LGAGVIEEVRVQRDKGFGFVRYSTHAEAALA 286
++ ++VG+L + T L F GA V+ + R KG+GFV++S +E A
Sbjct: 378 HS-IFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRA 554
Query: 287 IQMGN 291
+ N
Sbjct: 555 MSEMN 569
Score = 48.9 bits (115), Expect = 3e-06
Identities = 35/96 (36%), Positives = 50/96 (51%), Gaps = 1/96 (1%)
Frame = +2
Query: 226 PENNPQYTTVYVGNLGSEATQLDLHRHFHALGAGVIEEVRVQRDKGFGFVRYSTHAEAAL 285
PE + TT+YVGNL ++ +L ++F L G I V+V K GFV++ A A
Sbjct: 737 PEYDVNNTTIYVGNLDLNVSEEELKQNF--LQFGEIVSVKVHPGKACGFVQFGARASAEE 910
Query: 286 AIQMGNAQSYLCG-KIIKCSWGSKPTPPGTASNPLP 320
AIQ Q + G ++I+ SWG P TA +P
Sbjct: 911 AIQ--KMQGKILGQQVIRVSWGR----PQTARQDVP 1000
Score = 41.2 bits (95), Expect = 7e-04
Identities = 26/81 (32%), Positives = 41/81 (50%)
Frame = +2
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSSYGFIHYFDRRSAALAILTLNGRHL 89
YVGN+ V+E L++ F G + K+ + + GF+ + R SA AI + G+ L
Sbjct: 767 YVGNLDLNVSEEELKQNFLQFGEIVSVKVHPGK--ACGFVQFGARASAEEAIQKMQGKIL 940
Query: 90 FGQPIKVNWAYASGQREDTSG 110
Q I+V+W R+D G
Sbjct: 941 GQQVIRVSWGRPQTARQDVPG 1003
Score = 36.6 bits (83), Expect = 0.017
Identities = 22/76 (28%), Positives = 38/76 (49%), Gaps = 5/76 (6%)
Frame = +3
Query: 30 YVGNVHTQVTEPLLQEVF-AGTGPVEGCKLFRKEKSS----YGFIHYFDRRSAALAILTL 84
+VG++ VT+ LLQE F G V G K+ + YGF+ + D A+ +
Sbjct: 387 FVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEM 566
Query: 85 NGRHLFGQPIKVNWAY 100
NG + +P++++ Y
Sbjct: 567 NGVYCSTRPMRISCCY 614
>TC82618 similar to GP|9294614|dbj|BAB02953.1 DNA/RNA binding protein-like
{Arabidopsis thaliana}, partial (35%)
Length = 850
Score = 92.4 bits (228), Expect = 3e-19
Identities = 57/165 (34%), Positives = 90/165 (54%), Gaps = 10/165 (6%)
Frame = +1
Query: 21 SSLSVRKCRYVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKS----SYGFIHYFDRRS 76
SS + K +VG++H + E L FA TG + K+ R +++ YGF+ + +
Sbjct: 349 SSAADNKTLWVGDLHHWMDENYLHRCFASTGEIFSIKVIRNKQTCQTEGYGFVEFTSHGT 528
Query: 77 AALAILTLNGRHLFG--QPIKVNWA-YASG--QREDTSGHYNIFVGDLSPEVTDATLFAC 131
A + T G + QP ++NWA +++G +R D +IFVGDL+ +VTD L
Sbjct: 529 AEKVLQTYAGMLMPNTEQPFRLNWATFSTGDHKRSDNVPDLSIFVGDLAADVTDTMLLET 708
Query: 132 FS-VYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTG 175
FS Y S A+V++D TGRS+G+GFV F + A+N++ G
Sbjct: 709 FSDKYPSVKAAKVVFDANTGRSKGYGFVRFGDDGERSKALNEMNG 843
Score = 54.7 bits (130), Expect = 6e-08
Identities = 46/185 (24%), Positives = 79/185 (41%), Gaps = 7/185 (3%)
Frame = +1
Query: 114 IFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDL 173
++VGDL + + L CF+ +V+ +++T ++ G+GFV F S A+ +
Sbjct: 373 LWVGDLHHWMDENYLHRCFASTGEIFSIKVIRNKQTCQTEGYGFVEFTSHGTAEKVLQTY 552
Query: 174 TGKWLGSRQ--IRCNWATKGAGGIEEKQNSDSKSVVELTNGSSEDGKEISSNDVPENNPQ 231
G + + + R NWAT G D K S++VP+
Sbjct: 553 AGMLMPNTEQPFRLNWATFSTG----------------------DHKR--SDNVPD---- 648
Query: 232 YTTVYVGNLGSEATQLDLHRHFH-----ALGAGVIEEVRVQRDKGFGFVRYSTHAEAALA 286
+++VG+L ++ T L F A V+ + R KG+GFVR+ E + A
Sbjct: 649 -LSIFVGDLAADVTDTMLLETFSDKYPSVKAAKVVFDANTGRSKGYGFVRFGDDGERSKA 825
Query: 287 IQMGN 291
+ N
Sbjct: 826 LNEMN 840
>TC88545 similar to GP|13560783|gb|AAK30205.1 poly(A)-binding protein
{Daucus carota}, partial (35%)
Length = 866
Score = 86.3 bits (212), Expect = 2e-17
Identities = 69/257 (26%), Positives = 115/257 (43%), Gaps = 9/257 (3%)
Frame = +2
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFR----KEKSSYGFIHYFDRRSAALAILTLN 85
YVG++ VT+ L ++F G V ++ R ++ YG++++ + AA A+ LN
Sbjct: 188 YVGDLDHDVTDSQLYDLFNQIGQVVSVRICRDLASQQSLGYGYVNFSNPHDAAKAMDVLN 367
Query: 86 GRHLFGQPIKVNWAYASGQREDTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMW 145
L +PI++ +++ SG NIF+ +L + L+ FS++ + ++
Sbjct: 368 FTPLNNKPIRIMYSHRDPSVRK-SGAANIFIKNLDRAIDHKALYDTFSIFGNILSCKIAM 544
Query: 146 DQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGGIEEKQNSDSKS 205
D +G S+G+GFV F +++ AQSAI+ L G L + + G + KQ+ D+
Sbjct: 545 DA-SGLSKGYGFVQFENEESAQSAIDKLNGMLLNDKPVY-------VGHFQRKQDRDNAL 700
Query: 206 VVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHALGAGVIEEVR 265
+N ++ VYV NL T DL F G I
Sbjct: 701 ----------------------SNAKFNNVYVKNLSESVTDDDLKNTFGEY--GTITSAV 808
Query: 266 VQRD-----KGFGFVRY 277
V RD K FGFV +
Sbjct: 809 VMRDVDGKSKCFGFVNF 859
>TC76331 similar to GP|7673355|gb|AAF66823.1| poly(A)-binding protein
{Nicotiana tabacum}, partial (97%)
Length = 2581
Score = 84.7 bits (208), Expect = 5e-17
Identities = 72/273 (26%), Positives = 120/273 (43%), Gaps = 9/273 (3%)
Frame = +3
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFR----KEKSSYGFIHYFDRRSAALAILTLN 85
YVG++ VT+ L ++F G V ++ R + YG+++Y + + AA A+ LN
Sbjct: 453 YVGDLDMNVTDSQLYDLFNQLGQVVSVRVCRDLTTRRSLGYGYVNYSNPQDAARALDVLN 632
Query: 86 GRHLFGQPIKVNWAYASGQREDTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMW 145
L +PI++ +++ SG NIF+ +L + L FS + + +V
Sbjct: 633 FTPLNNRPIRIMYSHRDPSIR-KSGQGNIFIKNLDKAIDHKALHDTFSSFGNILSCKVAV 809
Query: 146 DQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGGIEEKQNSDSKS 205
D +G+S+G+GFV F +++ AQ AI L G L +Q+ G KQ +S
Sbjct: 810 D-GSGQSKGYGFVQFDTEEAAQKAIEKLNGMLLNDKQVY-------VGPFLRKQERESTG 965
Query: 206 VVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHALGAGVIEEVR 265
+ ++ V+V NL T +L + F G I
Sbjct: 966 ----------------------DRAKFNNVFVKNLSESTTDDELKKTFGEF--GTITSAV 1073
Query: 266 VQRD-----KGFGFVRYSTHAEAALAIQMGNAQ 293
V RD K FGFV + + +AA A++ N +
Sbjct: 1074VMRDGDGKSKCFGFVNFESTDDAARAVEALNGK 1172
Score = 61.2 bits (147), Expect = 7e-10
Identities = 44/172 (25%), Positives = 78/172 (44%), Gaps = 18/172 (10%)
Frame = +3
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRK---EKSSYGFIHYFDRRSAALAILTLNG 86
+V N+ T+ L++ F G + + R + +GF+++ AA A+ LNG
Sbjct: 990 FVKNLSESTTDDELKKTFGEFGTITSAVVMRDGDGKSKCFGFVNFESTDDAARAVEALNG 1169
Query: 87 RHLFGQPIKVNWAYASGQRE---------------DTSGHYNIFVGDLSPEVTDATLFAC 131
+ + + V A +RE D N++V +L + D L
Sbjct: 1170 KKIDDKEWYVGKAQKKSEREHELKIKFEQSMKEAADKYQGANLYVKNLDDSIADEKLKEL 1349
Query: 132 FSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQI 183
FS Y + + +VM D G SRG GFV+F + ++A A+ ++ GK + S+ +
Sbjct: 1350 FSSYGTITSCKVMRDPN-GVSRGSGFVAFSTPEEASRALLEMNGKMVASKPL 1502
Score = 40.0 bits (92), Expect = 0.002
Identities = 21/79 (26%), Positives = 38/79 (47%), Gaps = 3/79 (3%)
Frame = +3
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKS---SYGFIHYFDRRSAALAILTLNG 86
YV N+ + + L+E+F+ G + CK+ R GF+ + A+ A+L +NG
Sbjct: 1299 YVKNLDDSIADEKLKELFSSYGTITSCKVMRDPNGVSRGSGFVAFSTPEEASRALLEMNG 1478
Query: 87 RHLFGQPIKVNWAYASGQR 105
+ + +P+ V A R
Sbjct: 1479 KMVASKPLYVTLAQRKEDR 1535
>TC78142 similar to GP|7578881|gb|AAF64167.1| plastid-specific ribosomal
protein 2 precursor {Spinacia oleracea}, partial (65%)
Length = 819
Score = 73.6 bits (179), Expect = 1e-13
Identities = 56/187 (29%), Positives = 87/187 (45%), Gaps = 14/187 (7%)
Frame = +2
Query: 18 AAVSSLSVRKCRYVGNVHTQVTEPLLQEVFAGTGPVEGCKL----FRKEKSSYGFIHYFD 73
A V S ++RK YVGN+ V+ L+++ G VE ++ + K + F+
Sbjct: 230 ATVDSPALRKL-YVGNIPRTVSNDELEKIVQEHGAVEKAEVMYDKYSKRSRRFAFVTMKT 406
Query: 74 RRSAALAILTLNGRHLFGQPIKVNWA----------YASGQREDTSGHYNIFVGDLSPEV 123
A A LNG + G+ IKVN +G+ Y ++VG+L+ V
Sbjct: 407 VEDANAAAEKLNGTEIGGREIKVNITEKPLTTEGLPVQAGESTFVDSPYKVYVGNLAKNV 586
Query: 124 TDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQI 183
T +L FS + A+V T +S GFGFV+F S +D ++AI+ L ++I
Sbjct: 587 TSDSLKKFFSEKGNALSAKVSRAPGTSKSSGFGFVTFSSDEDVEAAISSFNNALLEGQKI 766
Query: 184 RCNWATK 190
R N A K
Sbjct: 767 RVNKA*K 787
Score = 64.7 bits (156), Expect = 6e-11
Identities = 54/189 (28%), Positives = 82/189 (42%), Gaps = 4/189 (2%)
Frame = +2
Query: 107 DTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDA 166
D+ ++VG++ V++ L + + A VM+D+ + RSR F FV+ ++ +DA
Sbjct: 239 DSPALRKLYVGNIPRTVSNDELEKIVQEHGAVEKAEVMYDKYSKRSRRFAFVTMKTVEDA 418
Query: 167 QSAINDLTGKWLGSRQIRCNWATKGAGGIEEKQNSDSKSVVELTNGSSEDGKEISSNDVP 226
+A L G +G R+I+ N I EK + T G E + D P
Sbjct: 419 NAAAEKLNGTEIGGREIKVN--------ITEKPLT--------TEGLPVQAGESTFVDSP 550
Query: 227 ENNPQYTTVYVGNLGSEATQLDLHRHF----HALGAGVIEEVRVQRDKGFGFVRYSTHAE 282
VYVGNL T L + F +AL A V + GFGFV +S+ +
Sbjct: 551 YK------VYVGNLAKNVTSDSLKKFFSEKGNALSAKVSRAPGTSKSSGFGFVTFSSDED 712
Query: 283 AALAIQMGN 291
AI N
Sbjct: 713 VEAAISSFN 739
>TC76341 similar to GP|7528270|gb|AAF63202.1| poly(A)-binding protein
{Cucumis sativus}, partial (90%)
Length = 2501
Score = 73.2 bits (178), Expect = 2e-13
Identities = 68/269 (25%), Positives = 114/269 (42%), Gaps = 11/269 (4%)
Frame = +3
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRK---EKSSYGFIHYFDRRSAALAILTLNG 86
++ N+ + L + F+ G + CK+ + YGF+ + SA AI LNG
Sbjct: 567 FIKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKLNG 746
Query: 87 RHLFGQPIKVNWAYASGQREDT---SGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARV 143
+ + + V R++ + N++V +LS T+ L F Y + + A +
Sbjct: 747 MLINDKQVFVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDDLKNEFGAYGTITSAVL 926
Query: 144 MWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGGIEEKQNSDS 203
M D GRS+ FGFV+F + +DA A+ L GK + ++ A K + +E +
Sbjct: 927 MRDAD-GRSKCFGFVNFENAEDAAKAVEALNGKKVDDKEWYVGKAQKKSEREQELKGRFE 1103
Query: 204 KSVVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHALGAGVIEE 263
++V E + Q +Y+ NL T L F G I
Sbjct: 1104QTVKESV----------------VDKFQGLNLYLKNLDDSITDEKLKEMFSEF--GTITS 1229
Query: 264 VRVQRD-----KGFGFVRYSTHAEAALAI 287
++ RD +G GFV +ST EA+ A+
Sbjct: 1230YKIMRDPNGVSRGSGFVAFSTPEEASRAL 1316
Score = 63.2 bits (152), Expect = 2e-10
Identities = 45/173 (26%), Positives = 80/173 (46%), Gaps = 19/173 (10%)
Frame = +3
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRK---EKSSYGFIHYFDRRSAALAILTLNG 86
YV N+ TE L+ F G + L R +GF+++ + AA A+ LNG
Sbjct: 840 YVKNLSESFTEDDLKNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEALNG 1019
Query: 87 RHLFGQPIKVNWAYASGQRE-DTSGHY---------------NIFVGDLSPEVTDATLFA 130
+ + + V A +RE + G + N+++ +L +TD L
Sbjct: 1020 KKVDDKEWYVGKAQKKSEREQELKGRFEQTVKESVVDKFQGLNLYLKNLDDSITDEKLKE 1199
Query: 131 CFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQI 183
FS + + + ++M D G SRG GFV+F + ++A A+ ++ GK + S+ +
Sbjct: 1200 MFSEFGTITSYKIMRDPN-GVSRGSGFVAFSTPEEASRALGEMNGKMIVSKPL 1355
Score = 61.2 bits (147), Expect = 7e-10
Identities = 57/242 (23%), Positives = 94/242 (38%), Gaps = 61/242 (25%)
Frame = +3
Query: 113 NIFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAIND 172
+++VGDL V D+ L+ F+ RV D T RS G+G+V+F + QDA A++
Sbjct: 297 SLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDV 476
Query: 173 LTGKWLGSRQIR--------------------------------------------CNWA 188
L + ++ IR C A
Sbjct: 477 LNFTPMNNKSIRVMYSHRDPSSRKSGTANIFIKNLDKTIDHKALHDTFSSFGQIMSCKIA 656
Query: 189 T------KGAGGIEEKQNSDSKSVVELTNGSSEDGKEI--------SSNDVPENNPQYTT 234
T KG G ++ + +++ ++ NG + K++ D + ++
Sbjct: 657 TDGSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVGHFLRKQDRDNVLSKTKFNN 836
Query: 235 VYVGNLGSEATQLDLHRHFHALG---AGVIEEVRVQRDKGFGFVRYSTHAEAALAIQMGN 291
VYV NL T+ DL F A G + V+ R K FGFV + +AA A++ N
Sbjct: 837 VYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEALN 1016
Query: 292 AQ 293
+
Sbjct: 1017GK 1022
Score = 32.7 bits (73), Expect = 0.25
Identities = 17/70 (24%), Positives = 35/70 (49%), Gaps = 3/70 (4%)
Frame = +3
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKS---SYGFIHYFDRRSAALAILTLNG 86
Y+ N+ +T+ L+E+F+ G + K+ R GF+ + A+ A+ +NG
Sbjct: 1152 YLKNLDDSITDEKLKEMFSEFGTITSYKIMRDPNGVSRGSGFVAFSTPEEASRALGEMNG 1331
Query: 87 RHLFGQPIKV 96
+ + +P+ V
Sbjct: 1332 KMIVSKPLYV 1361
>TC76790 similar to PIR|T06817|T06817 RNA-binding protein - garden pea,
partial (79%)
Length = 1141
Score = 70.5 bits (171), Expect = 1e-12
Identities = 52/171 (30%), Positives = 80/171 (46%), Gaps = 12/171 (7%)
Frame = +1
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSS----YGFIHYFDRRSAALAILTLN 85
+VGN V L +F G VE ++ ++ +GF+ A A+ N
Sbjct: 370 FVGNFPFDVDSEKLAMLFGQAGTVEIAEVIYNRQTDLSRGFGFVTMSTVEEAESAVEKFN 549
Query: 86 GRHLFGQPIKVNWAYASGQREDTSGH--------YNIFVGDLSPEVTDATLFACFSVYQS 137
G G+ + VN A G R + + I+V +L+ EV ++ L FS +
Sbjct: 550 GYDYNGRSLVVNKASPKGSRPERTERAPRTFEPVLRIYVANLAWEVDNSRLEQVFSEHGK 729
Query: 138 CSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWA 188
ARV++D++TGRSRGFGFV+ + + AI L G+ L R IR + A
Sbjct: 730 IVSARVVYDRETGRSRGFGFVTMSDETEMNDAIAALDGQSLEGRTIRVSVA 882
Score = 53.9 bits (128), Expect = 1e-07
Identities = 50/195 (25%), Positives = 78/195 (39%), Gaps = 4/195 (2%)
Frame = +1
Query: 114 IFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDL 173
+FVG+ +V L F + A V+++++T SRGFGFV+ + ++A+SA+
Sbjct: 367 LFVGNFPFDVDSEKLAMLFGQAGTVEIAEVIYNRQTDLSRGFGFVTMSTVEEAESAVEKF 546
Query: 174 TGKWLGSRQIRCNWATKGAGGIEEKQNSDSKSVVELTNGSSEDGKEISSNDVPENNPQYT 233
G R + N A+ E + + P
Sbjct: 547 NGYDYNGRSLVVNKASPKGSRPERTERA------------------------PRTFEPVL 654
Query: 234 TVYVGNLGSEATQLDLHR----HFHALGAGVIEEVRVQRDKGFGFVRYSTHAEAALAIQM 289
+YV NL E L + H + A V+ + R +GFGFV S E AI
Sbjct: 655 RIYVANLAWEVDNSRLEQVFSEHGKIVSARVVYDRETGRSRGFGFVTMSDETEMNDAIAA 834
Query: 290 GNAQSYLCGKIIKCS 304
+ QS L G+ I+ S
Sbjct: 835 LDGQS-LEGRTIRVS 876
>TC76789 similar to GP|15294254|gb|AAK95304.1 AT4g24770/F22K18_30
{Arabidopsis thaliana}, partial (62%)
Length = 1294
Score = 69.7 bits (169), Expect = 2e-12
Identities = 49/168 (29%), Positives = 81/168 (48%), Gaps = 9/168 (5%)
Frame = +3
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSS----YGFIHYFDRRSAALAILTLN 85
+VGN+ V L ++F +G VE ++ + +GF+ A+ +
Sbjct: 462 FVGNLPFDVDSEKLAQLFEQSGTVEIAEVIYNRDTDRSRGFGFVTMSTSEEVERAVNKFS 641
Query: 86 GRHLFGQPIKVNWAYASG-----QREDTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSD 140
G L G+ + VN A G Q + +VG+L +V +++L FS +
Sbjct: 642 GFELDGRLLTVNKAAPRGTPRLRQPRTFNSGLRAYVGNLPWDVDNSSLEQLFSEHGKVES 821
Query: 141 ARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWA 188
A+V++D++TGRSRGFGFV+ ++ + AI L G+ R IR N A
Sbjct: 822 AQVVYDRETGRSRGFGFVTMSNEAEMNDAIAALDGQSFNGRAIRVNVA 965
Score = 60.5 bits (145), Expect = 1e-09
Identities = 53/195 (27%), Positives = 81/195 (41%), Gaps = 5/195 (2%)
Frame = +3
Query: 106 EDTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQD 165
E+ S IFVG+L +V L F + A V++++ T RSRGFGFV+ + ++
Sbjct: 435 EEPSEDLKIFVGNLPFDVDSEKLAQLFEQSGTVEIAEVIYNRDTDRSRGFGFVTMSTSEE 614
Query: 166 AQSAINDLTGKWLGSRQIRCNWAT-KGAGGIEEKQNSDSKSVVELTNGSSEDGKEISSND 224
+ A+N +G L R + N A +G + + + +S
Sbjct: 615 VERAVNKFSGFELDGRLLTVNKAAPRGTPRLRQPRTFNSG-------------------- 734
Query: 225 VPENNPQYTTVYVGNLGSEATQLDLHRHFHALG----AGVIEEVRVQRDKGFGFVRYSTH 280
YVGNL + L + F G A V+ + R +GFGFV S
Sbjct: 735 --------LRAYVGNLPWDVDNSSLEQLFSEHGKVESAQVVYDRETGRSRGFGFVTMSNE 890
Query: 281 AEAALAIQMGNAQSY 295
AE AI + QS+
Sbjct: 891 AEMNDAIAALDGQSF 935
Score = 35.0 bits (79), Expect = 0.050
Identities = 27/93 (29%), Positives = 42/93 (45%), Gaps = 4/93 (4%)
Frame = +3
Query: 199 QNSDSKSVVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHALGA 258
+N + V T + + E++S + P + + ++VGNL + L + F G
Sbjct: 360 ENQADATWVGETESNENEEVEVASFEEPSEDLK---IFVGNLPFDVDSEKLAQLFEQSGT 530
Query: 259 GVIEEVRVQRD----KGFGFVRYSTHAEAALAI 287
I EV RD +GFGFV ST E A+
Sbjct: 531 VEIAEVIYNRDTDRSRGFGFVTMSTSEEVERAV 629
>TC76948 similar to SP|Q08935|ROC1_NICSY 29 kDa ribonucleoprotein A
chloroplast precursor (CP29A). [Wood tobacco] {Nicotiana
sylvestris}, partial (61%)
Length = 1374
Score = 68.2 bits (165), Expect = 5e-12
Identities = 53/190 (27%), Positives = 82/190 (42%), Gaps = 27/190 (14%)
Frame = +1
Query: 22 SLSVRKCRYVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSS----YGFIHYFDRRSA 77
S S + +VGN+ V L E+F G VE ++ + + +GF+
Sbjct: 325 SYSPNQRLFVGNLPFSVDSAQLAEIFENAGDVEMVEVIYDKSTGRSRGFGFVTMSSAAEV 504
Query: 78 ALAILTLNGRHLFGQPIKVNWAYASGQREDTS-----------------------GHYNI 114
A LNG + G+ ++VN R + S G +
Sbjct: 505 EAAAQQLNGYVVDGRELRVNAGPPPPPRSENSRFGENPRFGGDRPRGPPRGGSSDGDNRV 684
Query: 115 FVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLT 174
VG+L+ V + L + F +A+V++D+++GRSRGFGFV+F S + SAI L
Sbjct: 685 HVGNLAWGVDNLALESLFGEQGQVLEAKVIYDRESGRSRGFGFVTFSSADEVDSAIRTLD 864
Query: 175 GKWLGSRQIR 184
G L R IR
Sbjct: 865 GADLNGRAIR 894
Score = 63.5 bits (153), Expect = 1e-10
Identities = 54/184 (29%), Positives = 78/184 (42%), Gaps = 4/184 (2%)
Frame = +1
Query: 109 SGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQS 168
S + +FVG+L V A L F V++D+ TGRSRGFGFV+ S + ++
Sbjct: 331 SPNQRLFVGNLPFSVDSAQLAEIFENAGDVEMVEVIYDKSTGRSRGFGFVTMSSAAEVEA 510
Query: 169 AINDLTGKWLGSRQIRCNWATKGAGGIEEKQNSDSKSVVELTNGSSEDGKEISSNDVPEN 228
A L G + R++R N E + ++ G G + +N
Sbjct: 511 AAQQLNGYVVDGRELRVNAGPPPPPRSENSRFGENPRF----GGDRPRGPPRGGSSDGDN 678
Query: 229 NPQYTTVYVGNLGSEATQLDLHRHF----HALGAGVIEEVRVQRDKGFGFVRYSTHAEAA 284
V+VGNL L L F L A VI + R +GFGFV +S+ E
Sbjct: 679 R-----VHVGNLAWGVDNLALESLFGEQGQVLEAKVIYDRESGRSRGFGFVTFSSADEVD 843
Query: 285 LAIQ 288
AI+
Sbjct: 844 SAIR 855
Score = 38.1 bits (87), Expect = 0.006
Identities = 35/108 (32%), Positives = 49/108 (44%), Gaps = 9/108 (8%)
Frame = +1
Query: 213 SSEDGKEISSNDVPENNPQYTT---VYVGNLGSEATQLDLHRHFHALGAGVIEEVRVQRD 269
SSE +E + D ++ P Y+ ++VGNL L F AG +E V V D
Sbjct: 271 SSEFDQEEDTFDDGDDTPSYSPNQRLFVGNLPFSVDSAQLAEIFE--NAGDVEMVEVIYD 444
Query: 270 K------GFGFVRYSTHAEAALAIQMGNAQSYLCGKIIKCSWGSKPTP 311
K GFGFV S+ AE A Q N + G+ ++ + G P P
Sbjct: 445 KSTGRSRGFGFVTMSSAAEVEAAAQQLNGY-VVDGRELRVNAGPPPPP 585
>TC84024 similar to PIR|S77714|S77714 RNA-binding protein precursor 33K -
wood tobacco, partial (55%)
Length = 867
Score = 67.8 bits (164), Expect = 7e-12
Identities = 54/178 (30%), Positives = 77/178 (42%), Gaps = 6/178 (3%)
Frame = +3
Query: 114 IFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDL 173
++VG+L +T + L F+ + +++D+ T RSRGF FV+ +S ++A+ AI
Sbjct: 339 LYVGNLPFSMTSSQLADIFAEAGTVVSVEIVYDRLTDRSRGFAFVTMKSAEEAKEAIRMF 518
Query: 174 TGKWLGSRQIRCNWATKGAGGIEEKQNSDSKSVVELTNGSSEDGKEISSNDVPENNPQYT 233
G +G R + N+ GG L G D P
Sbjct: 519 DGSQVGGRSAKVNFPEVPKGG------------ERLVMGQKVRNSYRGFVDSPHK----- 647
Query: 234 TVYVGNLGSEATQLDLHRHFH----ALGAGVIEEVRVQRDKGFGFVRYST--HAEAAL 285
+Y GNLG DL F LGA +I E R +GFGFV + T H EAAL
Sbjct: 648 -IYAGNLGWGMNSQDLRDAFDEQPGLLGAKIIYERDNGRSRGFGFVTFETAEHLEAAL 818
Score = 58.5 bits (140), Expect = 4e-09
Identities = 43/175 (24%), Positives = 75/175 (42%), Gaps = 18/175 (10%)
Frame = +3
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKL----FRKEKSSYGFIHYFDRRSAALAILTLN 85
YVGN+ +T L ++FA G V ++ + F+ A AI +
Sbjct: 342 YVGNLPFSMTSSQLADIFAEAGTVVSVEIVYDRLTDRSRGFAFVTMKSAEEAKEAIRMFD 521
Query: 86 GRHLFGQPIKVNWA--------YASGQREDTS------GHYNIFVGDLSPEVTDATLFAC 131
G + G+ KVN+ GQ+ S + I+ G+L + L
Sbjct: 522 GSQVGGRSAKVNFPEVPKGGERLVMGQKVRNSYRGFVDSPHKIYAGNLGWGMNSQDLRDA 701
Query: 132 FSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCN 186
F A++++++ GRSRGFGFV+F + + ++A+N + + R +R N
Sbjct: 702 FDEQPGLLGAKIIYERDNGRSRGFGFVTFETAEHLEAALNAMNXVEVQGRPLRLN 866
>TC77756 similar to PIR|T06796|T06796 glycine-rich RNA-binding protein -
garden pea, partial (89%)
Length = 813
Score = 66.6 bits (161), Expect = 2e-11
Identities = 37/82 (45%), Positives = 55/82 (66%), Gaps = 1/82 (1%)
Frame = +3
Query: 114 IFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDL 173
+F+G LS V D +L F+ Y +ARV+ D++TGRSRGFGFV+F S++ A SA++ +
Sbjct: 192 LFIGGLSYNVDDQSLRDAFTTYGDVVEARVITDRETGRSRGFGFVNFTSEESATSALS-M 368
Query: 174 TGKWLGSRQIRCNWAT-KGAGG 194
G+ L R IR ++A + AGG
Sbjct: 369 DGQDLNGRNIRVSYANDRQAGG 434
Score = 30.4 bits (67), Expect = 1.2
Identities = 21/77 (27%), Positives = 34/77 (43%), Gaps = 4/77 (5%)
Frame = +3
Query: 235 VYVGNLGSEATQLDLHRHFHALG----AGVIEEVRVQRDKGFGFVRYSTHAEAALAIQMG 290
+++G L L F G A VI + R +GFGFV +++ A A+ M
Sbjct: 192 LFIGGLSYNVDDQSLRDAFTTYGDVVEARVITDRETGRSRGFGFVNFTSEESATSALSMD 371
Query: 291 NAQSYLCGKIIKCSWGS 307
L G+ I+ S+ +
Sbjct: 372 GQD--LNGRNIRVSYAN 416
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.132 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,766,191
Number of Sequences: 36976
Number of extensions: 165007
Number of successful extensions: 2728
Number of sequences better than 10.0: 171
Number of HSP's better than 10.0 without gapping: 1686
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2560
length of query: 392
length of database: 9,014,727
effective HSP length: 98
effective length of query: 294
effective length of database: 5,391,079
effective search space: 1584977226
effective search space used: 1584977226
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146708.15


