

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146706.7 + phase: 0 /pseudo
(617 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
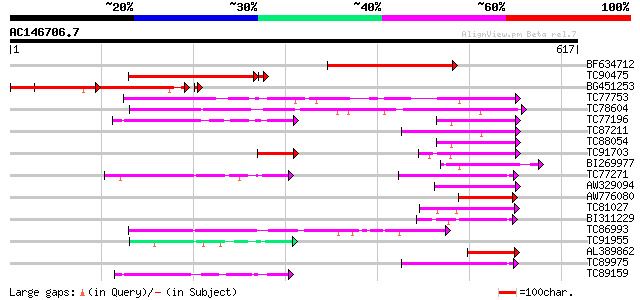
Score E
Sequences producing significant alignments: (bits) Value
BF634712 similar to PIR|T07943|T07 probable AMP-binding protein ... 279 2e-75
TC90475 similar to GP|17065456|gb|AAL32882.1 A6 anther-specific ... 218 2e-56
BG451253 similar to GP|17065456|gb| A6 anther-specific protein {... 204 2e-52
TC77753 similar to GP|19773582|gb|AAL98709.1 4-coumarate:coenzym... 112 3e-25
TC78604 similar to PIR|E84456|E84456 probable acyl-CoA synthetas... 97 2e-20
TC77196 similar to GP|17063848|gb|AAL35216.1 4-coumarate:CoA lig... 82 8e-16
TC87211 similar to GP|13516481|dbj|BAB40450. AMP-binding protein... 79 5e-15
TC88054 homologue to PIR|PQ0772|PQ0772 4-coumarate--CoA ligase (... 78 9e-15
TC91703 similar to GP|4038975|gb|AAC97600.1| 4-coumarate:CoA lig... 78 9e-15
BI269977 similar to GP|12039389|gb putative 4-coumarate CoA liga... 75 6e-14
TC77271 similar to PIR|T07929|T07929 probable long-chain-fatty-a... 72 5e-13
AW329094 similar to GP|20502991|gb Putative AMP-binding protein ... 72 8e-13
AW776080 similar to GP|20161027|db putative 4-coumarate-CoA liga... 71 1e-12
TC81027 similar to PIR|H85064|H85064 4-coumarate--CoA ligase-lik... 69 5e-12
BI311229 similar to GP|20502991|gb Putative AMP-binding protein ... 67 2e-11
TC86993 similar to GP|13516481|dbj|BAB40450. AMP-binding protein... 67 2e-11
TC91955 weakly similar to PIR|T27421|T27421 hypothetical protein... 63 4e-10
AL389862 similar to GP|23315116|gb Sequence 281 from patent US 6... 60 2e-09
TC89975 similar to PIR|G96530|G96530 probable acyl CoA synthetas... 60 3e-09
TC89159 weakly similar to PIR|H85064|H85064 4-coumarate--CoA lig... 58 1e-08
>BF634712 similar to PIR|T07943|T07 probable AMP-binding protein - rape,
partial (20%)
Length = 668
Score = 279 bits (714), Expect = 2e-75
Identities = 141/141 (100%), Positives = 141/141 (100%)
Frame = +2
Query: 347 VYESLYSGIQRQISTSSLVRKLVALTFIRVSLGYMECKRIYEGKCLTKNQKSPSYLYAML 406
VYESLYSGIQRQISTSSLVRKLVALTFIRVSLGYMECKRIYEGKCLTKNQKSPSYLYAML
Sbjct: 20 VYESLYSGIQRQISTSSLVRKLVALTFIRVSLGYMECKRIYEGKCLTKNQKSPSYLYAML 199
Query: 407 DWLGARIIATILFPIHMLAKKLVYSKIHSAIGFSKAGISGGGSLPSHVDRFFEAIGVTLQ 466
DWLGARIIATILFPIHMLAKKLVYSKIHSAIGFSKAGISGGGSLPSHVDRFFEAIGVTLQ
Sbjct: 200 DWLGARIIATILFPIHMLAKKLVYSKIHSAIGFSKAGISGGGSLPSHVDRFFEAIGVTLQ 379
Query: 467 NGYGLTETSPVIAARRLSCNV 487
NGYGLTETSPVIAARRLSCNV
Sbjct: 380 NGYGLTETSPVIAARRLSCNV 442
>TC90475 similar to GP|17065456|gb|AAL32882.1 A6 anther-specific protein
{Arabidopsis thaliana}, partial (15%)
Length = 761
Score = 218 bits (554), Expect(2) = 2e-56
Identities = 109/141 (77%), Positives = 123/141 (86%)
Frame = +2
Query: 130 MTYKQLEDAILDFAEGLRVIGVSPNEKIALFADNSCRWLVADQGMMATGAINVVRGSRSS 189
+T QL+DAIL+FAEGLRVIGV P+EKIALFADNSCRWLVADQGMMA GAINVVRGSRSS
Sbjct: 185 LTTFQLKDAILNFAEGLRVIGVKPDEKIALFADNSCRWLVADQGMMAIGAINVVRGSRSS 364
Query: 190 IEELLQIYNHSESIALAVDNPEMFNRIAKAFDLKASMRFVILLWGEKSCLVNEGSKEVPI 249
IEELLQIYNHSES+ALAVDNPEM+N+IAK F LK S+RF+ILLWGEKS LVNE K VPI
Sbjct: 365 IEELLQIYNHSESVALAVDNPEMYNQIAKPFYLKTSIRFIILLWGEKSSLVNEADKGVPI 544
Query: 250 FTFTEIMHLGRGSRRLFESHD 270
+TF E+++ GR SRR + D
Sbjct: 545 YTFMEVINFGRQSRRALHTSD 607
Score = 20.4 bits (41), Expect(2) = 2e-56
Identities = 7/11 (63%), Positives = 9/11 (81%)
Frame = +3
Query: 271 ARKHYVFEAIK 281
A +HY+FE IK
Sbjct: 726 AGEHYIFETIK 758
>BG451253 similar to GP|17065456|gb| A6 anther-specific protein {Arabidopsis
thaliana}, partial (12%)
Length = 686
Score = 204 bits (518), Expect(2) = 2e-52
Identities = 117/176 (66%), Positives = 124/176 (69%), Gaps = 8/176 (4%)
Frame = +1
Query: 28 KCQRFPSLPNATTILLQPSSPILFLSSFLATASPLLSTFLELVSFANLNPRFSPLLESSL 87
KC + S + S P+LF T+ L T + S + + RFSPLLESSL
Sbjct: 151 KCNNYTS-----STFFSYSLPLLFPRHSFTTSINLSRTRIFCQSQSKIR-RFSPLLESSL 312
Query: 88 LSGNDGVVSDEWKTVPDIWRSSAEKYGDKIALIDPYHDPPSTMTYKQLEDAILDFAEGLR 147
LSGNDGVVSDEWKTVPDIWRSSAEKYGDKIALIDPYHDPPSTMTYKQLEDAILDFAEGLR
Sbjct: 313 LSGNDGVVSDEWKTVPDIWRSSAEKYGDKIALIDPYHDPPSTMTYKQLEDAILDFAEGLR 492
Query: 148 VIGVSPNEKIALFADNSCRWLVADQ--------GMMATGAINVVRGSRSSIEELLQ 195
VIGVSPNEKIALFADNSCRWLVADQ M G RSSIEELLQ
Sbjct: 493 VIGVSPNEKIALFADNSCRWLVADQV*V*WQPEQSMLLGV------QRSSIEELLQ 642
Score = 160 bits (405), Expect = 1e-39
Identities = 84/110 (76%), Positives = 89/110 (80%), Gaps = 12/110 (10%)
Frame = +2
Query: 1 MVIRSSTHLFKSNHHFFFFFFFFFYKVKCQRFPSLPNATTILLQPSSPILFLSSFLATAS 60
MVIRSSTHLFKSNHHFFFFFFFFFYKVKCQRFPSLPNATTILLQPSSPILFLSSFLATAS
Sbjct: 44 MVIRSSTHLFKSNHHFFFFFFFFFYKVKCQRFPSLPNATTILLQPSSPILFLSSFLATAS 223
Query: 61 PLLSTFLELVSFANLNPR------------FSPLLESSLLSGNDGVVSDE 98
PLLSTFLELVSFANLNPR F ++ SS +SG ++ E
Sbjct: 224 PLLSTFLELVSFANLNPR*EGSPLY*KVHCFLVMMVSSRMSGKQFLIFGE 373
Score = 40.4 bits (93), Expect = 0.002
Identities = 20/31 (64%), Positives = 23/31 (73%)
Frame = +3
Query: 173 GMMATGAINVVRGSRSSIEELLQIYNHSESI 203
GMMATGAINVVRGS+ IYNHSE++
Sbjct: 573 GMMATGAINVVRGSKVIN*RTAAIYNHSEAL 665
Score = 21.2 bits (43), Expect(2) = 2e-52
Identities = 9/9 (100%), Positives = 9/9 (100%)
Frame = +2
Query: 202 SIALAVDNP 210
SIALAVDNP
Sbjct: 659 SIALAVDNP 685
>TC77753 similar to GP|19773582|gb|AAL98709.1 4-coumarate:coenzyme A ligase
{Glycine max}, partial (94%)
Length = 2034
Score = 112 bits (281), Expect = 3e-25
Identities = 108/458 (23%), Positives = 187/458 (40%), Gaps = 27/458 (5%)
Frame = +3
Query: 125 DPPSTMTYKQLEDAILDFAEGLRVIGVSPNEKIALFADNSCRWLVADQGMMATGAINVVR 184
D T+TY + + A GL +G+ + I + NS ++ + G GA+
Sbjct: 207 DTGETLTYSDVHLTVRKIAAGLNTLGIQQGDVIMIVLRNSPQFALTFLGASFRGAVITTA 386
Query: 185 GSRSSIEELLQIYNHSESIALAVDNPEM--FNRIAKAFDLKASMRFVILLWGEKSCLVNE 242
+ EL + ++S + + + N AK D+K C+ +
Sbjct: 387 NPFYTSSELAKQATATKSKLIITQSVYLNKINDFAKLIDIKIV------------CIDSS 530
Query: 243 GSKEVPIFTFTEIMHLGRGSRRLFESHDARKHYVFEAIKSDDIATLVYTSGTTGNPKGVM 302
++ + F+ + + E + + I +D+ L ++SGT+G PKGVM
Sbjct: 531 PEEDENVVDFSVLTNADEN-----ELPEVK-------INPNDVVALPFSSGTSGLPKGVM 674
Query: 303 LTHQNLL------------HQISKYAASLACI*KSL*VFHFF---------MRC*PSIYN 341
LTH+NL+ HQ + Y L C+ L +FH + +R ++
Sbjct: 675 LTHENLVTTISQLVDGENPHQYTNYEDVLLCV---LPMFHIYALNSILLCGIRSGAAVLI 845
Query: 342 CEKLEVYESLYSGIQRQISTSSLVRKLVALTFIRVSLGYMECKRIYEGKCLTKNQKSPSY 401
EK E+ + L + +++ +S V +V L K+ +S Y
Sbjct: 846 VEKFEITKVLELIEKYKVTVASFVPPIVL--------------------ALVKSGESMRY 965
Query: 402 LYAMLDWLGARIIATILFPIHMLAKKLVYSKIHSAIGFSKAGISGGGSLPSHVDRFFEAI 461
D R++ T P+ M ++ V ++ +
Sbjct: 966 -----DLSSIRVMITGAAPMGMELEQAVKDRLPRTV------------------------ 1058
Query: 462 GVTLQNGYGLTETSPVIAARRLSCNVI----GSVGHPLKHTEFKVVDSETGEVLPPGYKG 517
L GYG+TE P+ + + G+ G +++ E K+VD+ETG LP G
Sbjct: 1059---LGQGYGMTEAGPLSISLAFAKEPFKTKPGACGTVVRNAEMKIVDTETGASLPRNKAG 1229
Query: 518 ILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGWI 555
+ +RG ++MKGY +P AT + IDK+GWL+TGDIG I
Sbjct: 1230EICIRGTKVMKGYLNDPEATKRTIDKEGWLHTGDIGLI 1343
>TC78604 similar to PIR|E84456|E84456 probable acyl-CoA synthetase
[imported] - Arabidopsis thaliana, partial (91%)
Length = 2573
Score = 96.7 bits (239), Expect = 2e-20
Identities = 116/469 (24%), Positives = 197/469 (41%), Gaps = 37/469 (7%)
Frame = +2
Query: 131 TYKQLEDAILDFAEGLRVIGVSPNEKIALFADNSCRWLVADQGMMATGAINVVRGSRSSI 190
TY ++ + +FA GL +G + +A+F+D W +A QG V +
Sbjct: 488 TYGEVFARVSNFASGLLKLGHDIDSHVAIFSDTRAEWFIALQGCFRQNITVVTIYASLGE 667
Query: 191 EELLQIYNHSESIALAVDNPEMFNRIAKAFDLKASMRFVILLWGE--KSCLVNEG-SKEV 247
+ L+ N ++ L D ++ N++ S++ +I + + +EG S
Sbjct: 668 DALIHSLNETQVSTLICD-VKLLNKLDAIRSKLTSLQNIIYFEDDSKEEHTFSEGLSSNC 844
Query: 248 PIFTFTEIMHLGRGSRRLFESHDARKHYVFEAIKSDD-IATLVYTSGTTGNPKGVMLTHQ 306
I +F E+ LG+ S V ++ S + +A ++YTSG+TG PKGVM+TH
Sbjct: 845 KIASFDEVEKLGKESP------------VEPSLPSKNAVAVVMYTSGSTGLPKGVMITHG 988
Query: 307 NLLHQISKYAASLACI*KSL*VFHFFMRC*PSIYNCEKLEVYESLYSGI-------QRQI 359
N++ A++ + +L ++ P + E L +G+
Sbjct: 989 NIVAT----TAAVMTVIPNLGSKDVYLAYLPLAHGFEMAAESVMLAAGVAIGYGSPMTLT 1156
Query: 360 STSSLVRK-------------LVALTFIRVSLGYMECKRIYEGKCLTKNQKSPSYLYAML 406
TS+ V+K L A+ I + K++ E L KN +Y +
Sbjct: 1157DTSNKVKKGTKGDVTVLKPTLLTAVPAIIDRIRDGVVKKVEEKGGLAKNLFQIAYKRRLA 1336
Query: 407 ----DWLGARIIATILFPIHMLAKKLVYSKIHSAIGFS-KAGISGGGSLPSHVDRFFE-A 460
WLGA + +++ +++ KI + +G + + + GG L +F
Sbjct: 1337AVKGSWLGAWGVEKLVWDT------IIFKKIRTVLGGNLRFMLCGGAPLSGDSQQFINIC 1498
Query: 461 IGVTLQNGYGLTETSPVIAARRLSCNVIGSVGHPLKHTEFKVVDSETGEVL---PPGYKG 517
+G + GYGLTET A +G VG PL K+V E G L P +G
Sbjct: 1499VGAPIGQGYGLTETFAGAAFSEADDYSVGRVGPPLPCCYIKLVSWEEGGYLSSDKPMPRG 1678
Query: 518 ILKVRGPQLMKGYYKNPSATNQA--IDKDG--WLNTGDIGWIAAYHSSG 562
+ V G + GY+KN T++ +D+ G W TGDIG +H G
Sbjct: 1679EVVVGGFSVTAGYFKNQDKTDEVFKVDEKGVRWFYTGDIG---RFHPDG 1816
>TC77196 similar to GP|17063848|gb|AAL35216.1 4-coumarate:CoA ligase {Amorpha
fruticosa}, partial (93%)
Length = 1924
Score = 81.6 bits (200), Expect = 8e-16
Identities = 41/96 (42%), Positives = 57/96 (58%), Gaps = 5/96 (5%)
Frame = +3
Query: 465 LQNGYGLTETSPVIA-----ARRLSCNVIGSVGHPLKHTEFKVVDSETGEVLPPGYKGIL 519
L GYG+TE PV+ A+ G+ G +++ E K+VD + LP G +
Sbjct: 1101 LGQGYGMTEAGPVLTMCLSFAKEPIDVKSGACGTVVRNAEMKIVDPQNDSSLPRNQPGEI 1280
Query: 520 KVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGWI 555
+RG Q+MKGY NP AT + IDK+GWL+TGDIG+I
Sbjct: 1281 CIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIGFI 1388
Score = 50.1 bits (118), Expect = 3e-06
Identities = 51/203 (25%), Positives = 84/203 (41%)
Frame = +1
Query: 112 KYGDKIALIDPYHDPPSTMTYKQLEDAILDFAEGLRVIGVSPNEKIALFADNSCRWLVAD 171
KYG + LI+ TY ++ A GL +G+ + I + N ++ A
Sbjct: 220 KYGSRPCLINA--PTAEIYTYYDVQLTAQKVASGLNKLGIQQGDVIMVLLPNCPEFVFAF 393
Query: 172 QGMMATGAINVVRGSRSSIEELLQIYNHSESIALAVDNPEMFNRIAKAFDLKASMRFVIL 231
G GAI + E+ + S + L A +D + V L
Sbjct: 394 LGASFRGAIMTAANPFFTSAEIAKQAKASNTKLLVTQ--------ACYYDKVKDLENVKL 549
Query: 232 LWGEKSCLVNEGSKEVPIFTFTEIMHLGRGSRRLFESHDARKHYVFEAIKSDDIATLVYT 291
++ V+ +E F+E++ + E + + + IK DD+ L Y+
Sbjct: 550 VF------VDSSPEEDNHMHFSELIQADQN-----EMEEVKVN-----IKPDDVVALPYS 681
Query: 292 SGTTGNPKGVMLTHQNLLHQISK 314
SGTTG PKGVMLTH+ L+ I++
Sbjct: 682 SGTTGLPKGVMLTHKGLVTSIAQ 750
>TC87211 similar to GP|13516481|dbj|BAB40450. AMP-binding protein
{Arabidopsis thaliana}, partial (52%)
Length = 1338
Score = 79.0 bits (193), Expect = 5e-15
Identities = 51/136 (37%), Positives = 74/136 (53%), Gaps = 6/136 (4%)
Frame = +3
Query: 427 KLVYSKIHSAIGFS-KAGISGGGSLPSHVDRFFE-AIGVTLQNGYGLTETSPVIAARRLS 484
+LV++KI +G + +SG L V F + G + GYG+TET+ VI+
Sbjct: 222 RLVFNKIKEKLGGRVRLMVSGASPLSPDVMEFLKICFGGRVTEGYGMTETTCVISCIDNG 401
Query: 485 CNVIGSVGHPLKHTEFKVVDSETGEVLP---PGYKGILKVRGPQLMKGYYKNPSATNQAI 541
+ G VG P E K+VD P +G + VRGP + +GYYK+ + T + I
Sbjct: 402 DRLGGHVGSPNPACEIKLVDVPEMNYTSDDQPNPRGEICVRGPIIFQGYYKDEAQTREVI 581
Query: 542 DKDGWLNTGDIG-WIA 556
D++GWL+TGDIG WIA
Sbjct: 582 DEEGWLHTGDIGTWIA 629
>TC88054 homologue to PIR|PQ0772|PQ0772 4-coumarate--CoA ligase (EC
6.2.1.12) (clone GM4CL1B) - soybean (fragment), partial
(62%)
Length = 1055
Score = 78.2 bits (191), Expect = 9e-15
Identities = 40/96 (41%), Positives = 58/96 (59%), Gaps = 5/96 (5%)
Frame = +1
Query: 465 LQNGYGLTETSPVIA-----ARRLSCNVIGSVGHPLKHTEFKVVDSETGEVLPPGYKGIL 519
L GYG+TE PV++ A+ GS G +++ E KV+D ETG L G +
Sbjct: 169 LGQGYGMTEAGPVLSMSLGFAKNPFPTSSGSCGTVVRNAELKVLDPETGRSLGYNQPGEI 348
Query: 520 KVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGWI 555
+RG Q+MKGY + +AT ID++GWL+TGD+G+I
Sbjct: 349 CIRGQQIMKGYLNDENATKTTIDEEGWLHTGDVGYI 456
>TC91703 similar to GP|4038975|gb|AAC97600.1| 4-coumarate:CoA ligase
isoenzyme 2 {Glycine max}, partial (48%)
Length = 828
Score = 78.2 bits (191), Expect = 9e-15
Identities = 49/121 (40%), Positives = 68/121 (55%), Gaps = 10/121 (8%)
Frame = +1
Query: 445 SGGGSLPSHVD-----RFFEAIGVTLQNGYGLTETSPVI----AARRLSCNVIGSV-GHP 494
SGG L ++ +F AI L GY +TE PV+ A + NV S G
Sbjct: 421 SGGAPLGKELEDTVRTKFPNAI---LGQGYRMTEAGPVLTMSLAFAKEPLNVKASACGTV 591
Query: 495 LKHTEFKVVDSETGEVLPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGW 554
+++ E K+VD +TG+ LP G + +RG Q+MKGY + AT + IDK+GWL TGDIG+
Sbjct: 592 VRNAEMKIVDPDTGKSLPRNQSGEICIRGDQIMKGYLNDLEATERTIDKEGWLYTGDIGY 771
Query: 555 I 555
I
Sbjct: 772 I 774
Score = 47.4 bits (111), Expect = 2e-05
Identities = 22/45 (48%), Positives = 29/45 (63%)
Frame = +1
Query: 270 DARKHYVFEAIKSDDIATLVYTSGTTGNPKGVMLTHQNLLHQISK 314
D H I+ DD+ L Y+SGTTG PKGVMLTH+ L+ I++
Sbjct: 4 DENDHLPDAKIQPDDVVALPYSSGTTGLPKGVMLTHKGLVSSIAQ 138
>BI269977 similar to GP|12039389|gb putative 4-coumarate CoA ligase {Oryza
sativa}, partial (28%)
Length = 520
Score = 75.5 bits (184), Expect = 6e-14
Identities = 46/116 (39%), Positives = 62/116 (52%), Gaps = 3/116 (2%)
Frame = +1
Query: 469 YGLTETSPVIAARRLSCNVI---GSVGHPLKHTEFKVVDSETGEVLPPGYKGILKVRGPQ 525
YG+TE S + R + + S+G + E KVVD ++G LPPG G L +RGP
Sbjct: 1 YGMTE-SGAVGTRGFNTDKFHNYSSLGLLAPNIEAKVVDWKSGTFLPPGLSGELWLRGPS 177
Query: 526 LMKGYYKNPSATNQAIDKDGWLNTGDIGWIAAYHSSGRSRNCGGVIVVEGRAKDTI 581
+MKGY N AT IDKDGW++TGDI + + G + + GR KD I
Sbjct: 178 IMKGYLNNEEATMSTIDKDGWIHTGDIVYF----------DQDGYLYMSGRLKDII 315
>TC77271 similar to PIR|T07929|T07929 probable long-chain-fatty-acid--CoA
ligase (EC 6.2.1.3) isoform 2 - rape, partial (94%)
Length = 2486
Score = 72.4 bits (176), Expect = 5e-13
Identities = 55/216 (25%), Positives = 100/216 (45%), Gaps = 10/216 (4%)
Frame = +2
Query: 104 DIWRSSAEKYGDKIAL-----IDPYHDPPSTMTYKQLEDAILDFAEGLRVIGVSPNEKIA 158
D++R S EKY + L +D H TYK++ D ++ +R G K
Sbjct: 305 DVFRLSVEKYPNNPMLGSREIVDGKHGKYKWQTYKEVYDMVIKVGNSIRSCGYGEGVKCG 484
Query: 159 LFADNSCRWLVADQGMMATGAINVVRGSRSSIEELLQIYNHSESIALAVDNPEMFNRIAK 218
++ NS W+++ + A G V + I +H+E +++A + + K
Sbjct: 485 IYGANSAEWIMSMEACNAHGLHCVPLYDTLGSGAIEFIISHAE-VSIAFAEEKKIPELLK 661
Query: 219 AFDLKASMRFVILLWGEKSCLVNEGSKEVP-----IFTFTEIMHLGRGSRRLFESHDARK 273
F I+ +G+ + E +EV I+++TE + LG+ SH
Sbjct: 662 TFPNATKYLKTIVSFGK---VTPEQKQEVEKFGLAIYSWTEFLQLGK-------SHS--- 802
Query: 274 HYVFEAIKSDDIATLVYTSGTTGNPKGVMLTHQNLL 309
+ K DI T++YTSGTTG+PKGV++++++++
Sbjct: 803 -FDLPVKKRSDICTIMYTSGTTGDPKGVLISNESII 907
Score = 61.2 bits (147), Expect = 1e-09
Identities = 41/135 (30%), Positives = 68/135 (50%), Gaps = 5/135 (3%)
Frame = +1
Query: 424 LAKKLVYSKIHSAIGFS-KAGISGGGSLPSHVDRFFEAIGVT-LQNGYGLTET-SPVIAA 480
L K+V+ K+ +G S + +SG L HV+ + + + GYGLTET + +
Sbjct: 1273 LLDKIVFDKVKQGLGGSVRLILSGAAPLSLHVESYLRVVTCAHVLQGYGLTETCAGTFVS 1452
Query: 481 RRLSCNVIGSVGHPLKHTE--FKVVDSETGEVLPPGYKGILKVRGPQLMKGYYKNPSATN 538
+++G+VG P+ + + + V + L +G + V+G L GYYK T
Sbjct: 1453 LPNELDMLGTVGPPVPNVDACLESVPEMGYDALASTPRGEICVKGDTLFSGYYKREDLTK 1632
Query: 539 QAIDKDGWLNTGDIG 553
+ + DGW +TGDIG
Sbjct: 1633 EVL-VDGWFHTGDIG 1674
>AW329094 similar to GP|20502991|gb Putative AMP-binding protein {Oryza
sativa (japonica cultivar-group)}, partial (34%)
Length = 595
Score = 71.6 bits (174), Expect = 8e-13
Identities = 39/95 (41%), Positives = 53/95 (55%), Gaps = 2/95 (2%)
Frame = +2
Query: 463 VTLQNGYGLTETSPVIAARRL--SCNVIGSVGHPLKHTEFKVVDSETGEVLPPGYKGILK 520
V++ GYGLTE+S + A+ G+ G TE +VD+ET + LP G L
Sbjct: 59 VSIFQGYGLTESSGIGASTESLEESRKYGTAGLVSASTEAMIVDTETAQPLPVNRTGELW 238
Query: 521 VRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGWI 555
+RGP MKGY+ N AT I +GWL TGD+ +I
Sbjct: 239 LRGPTTMKGYFSNEEATRSTITPEGWLKTGDVCYI 343
>AW776080 similar to GP|20161027|db putative 4-coumarate-CoA ligase {Oryza
sativa (japonica cultivar-group)}, partial (28%)
Length = 625
Score = 71.2 bits (173), Expect = 1e-12
Identities = 33/64 (51%), Positives = 43/64 (66%)
Frame = +2
Query: 489 GSVGHPLKHTEFKVVDSETGEVLPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLN 548
GSVG ++ E K+VD TGE L PG KG L +RGP +MKGY + AT + +D +GWL
Sbjct: 65 GSVGRLAENMEAKIVDPVTGEALSPGQKGELWLRGPTIMKGYVGDDKATVETLDSEGWLK 244
Query: 549 TGDI 552
TGD+
Sbjct: 245 TGDL 256
>TC81027 similar to PIR|H85064|H85064 4-coumarate--CoA ligase-like protein
[imported] - Arabidopsis thaliana, partial (28%)
Length = 1283
Score = 68.9 bits (167), Expect = 5e-12
Identities = 38/116 (32%), Positives = 61/116 (51%), Gaps = 8/116 (6%)
Frame = +1
Query: 447 GGSLPSHVDRFFEAIGVT----LQNGYGLTETSPVIAARRLS----CNVIGSVGHPLKHT 498
GG+ P D E + + GYG+TE +++ +V GS G +
Sbjct: 343 GGAAPLGKDLMQECTKILPHVHVIQGYGMTEACGLVSIENPKEGSLISVSGSTGTLIPSV 522
Query: 499 EFKVVDSETGEVLPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGW 554
E ++++ T + LPP G + +RGP +M+GY+ NP AT AI+ GW+ TGD+G+
Sbjct: 523 ESRIINLATLKPLPPNQLGEIWLRGPTIMQGYFNNPEATKLAINDQGWMITGDLGY 690
Score = 29.3 bits (64), Expect = 4.7
Identities = 12/15 (80%), Positives = 13/15 (86%)
Frame = +1
Query: 295 TGNPKGVMLTHQNLL 309
TG KGVMLTHQNL+
Sbjct: 1 TGRSKGVMLTHQNLI 45
>BI311229 similar to GP|20502991|gb Putative AMP-binding protein {Oryza
sativa (japonica cultivar-group)}, partial (29%)
Length = 693
Score = 67.4 bits (163), Expect = 2e-11
Identities = 43/114 (37%), Positives = 63/114 (54%), Gaps = 4/114 (3%)
Frame = +3
Query: 443 GISGGGSLPSHVDRFFEAIGVTLQNGYGLTETSP----VIAARRLSCNVIGSVGHPLKHT 498
G G G+ + + +F A ++ GYGLTE++ ++ S G+ G +
Sbjct: 3 GSVGKGNFVAFMAKFPHA---SIIQGYGLTESTAGVIRIVGPEEASRG--GTTGKLVSGM 167
Query: 499 EFKVVDSETGEVLPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDI 552
E K+V+ TGE + PG +G L VRGP +MKGY +P AT+ + DGWL TGDI
Sbjct: 168 EAKIVNPNTGEAMSPGEQGELWVRGPPIMKGYVGDPVATSVTL-VDGWLRTGDI 326
>TC86993 similar to GP|13516481|dbj|BAB40450. AMP-binding protein
{Arabidopsis thaliana}, partial (65%)
Length = 1523
Score = 67.0 bits (162), Expect = 2e-11
Identities = 90/364 (24%), Positives = 149/364 (40%), Gaps = 14/364 (3%)
Frame = +2
Query: 130 MTYKQLEDAILDFAEGLRVIGVSPNEKIALFADNSCRWLVADQGMMATGAINVVRGSRSS 189
MTY + A GL G+ I L+ N WL+ D A I+V
Sbjct: 416 MTYGEAGTARSAIGSGLIHYGIPKGAGIGLYFINRPEWLIVDHACSAYSYISVPLYDTLG 595
Query: 190 IEELLQIYNHSESIALAVDNPEMFNRIAKAFDLKASMRFVILLWGEKSCLVNEGSKE-VP 248
+ + I NH+ + + + N + ++R ++++ G + + S + V
Sbjct: 596 PDAVKYIVNHA-LVQVIFCVSQTLNSLLSYLSEIPTVRLIVVVGGIDDQIPSLPSSDGVQ 772
Query: 249 IFTFTEIMHLGRGSRRLFESHDARKHYVFEAIKSDDIATLVYTSGTTGNPKGVMLTHQNL 308
I ++T++ GR + + F K +D+AT+ YTSGTTG PKG +LTH+N
Sbjct: 773 IISYTKLFSQGRSNLQPFCPP-----------KPEDVATICYTSGTTGTPKGAVLTHENF 919
Query: 309 LHQISKYAASLACI*KSL*VFHFFMRC*PSIYNCEKLEVYESLYSGIQ---RQISTSSLV 365
+ ++ A I + ++ P + E+ ++Y G+ Q L+
Sbjct: 920 IANVAG-----ATIDEKFNPSDVYISYLPLAHIYERANQVMTVYFGMAVGFYQGDNLKLM 1084
Query: 366 RKLVAL---TFIRVSLGYMECKRIYEGKCLTKNQKSPSYLYAMLDWLGARIIATILFPIH 422
L AL F V Y RIY G + K+ L L A A +H
Sbjct: 1085DDLAALRPTVFCSVPRLY---NRIYAG--IINAVKTSGGLKERL--FNAAYNAKRQALLH 1243
Query: 423 -----MLAKKLVYSKIHSAIGFS-KAGISGGGSLPSHVDRFFE-AIGVTLQNGYGLTETS 475
+ +LV++KI +G + +SG L V F + G + GYG+TET+
Sbjct: 1244GKNPSPMWDRLVFNKIKEKLGGRVRLMVSGASPLSPDVMEFLKICFGGRVTEGYGMTETT 1423
Query: 476 PVIA 479
VI+
Sbjct: 1424CVIS 1435
>TC91955 weakly similar to PIR|T27421|T27421 hypothetical protein Y76A2B.3 -
Caenorhabditis elegans, partial (7%)
Length = 894
Score = 62.8 bits (151), Expect = 4e-10
Identities = 58/221 (26%), Positives = 88/221 (39%), Gaps = 38/221 (17%)
Frame = +2
Query: 131 TYKQLEDAILDFAEGLRVIGVSPNEK----------------IALFADNSCRWLVADQGM 174
TYKQ+ + I +F GL + + N+ + ++A+N W + DQ
Sbjct: 197 TYKQIAERIDNFGSGLLYLNENTNKNPKVKKNPKNFKDKQWTVGIYANNRPEWFITDQAN 376
Query: 175 MATGAINVVRGSRSSIEELLQIYNHSESIALAVDN----PEMFNRIAKAFDLKASMRF-- 228
A I V E + + NH+E I + V P + +K +LK +
Sbjct: 377 CAYNLITVALYDTXGPETVEFVINHAE-IPIVVTGASRIPGLIQLASKVPNLKVIISMDE 553
Query: 229 ----------------VILLWGEKSCLVNEGSKEVPIFTFTEIMHLGRGSRRLFESHDAR 272
V+ W E K + + +F+E+ LG + R
Sbjct: 554 LEDDSPVPFGSTTTGKVLKAWAE--------DKGIVLLSFSEVEKLG--------NQYPR 685
Query: 273 KHYVFEAIKSDDIATLVYTSGTTGNPKGVMLTHQNLLHQIS 313
KH D+A + YTSGTTG PKGVMLTH+N + IS
Sbjct: 686 KH---NPPSPKDLACICYTSGTTGVPKGVMLTHRNFVAAIS 799
>AL389862 similar to GP|23315116|gb Sequence 281 from patent US 6410718,
partial (90%)
Length = 535
Score = 60.1 bits (144), Expect = 2e-09
Identities = 25/56 (44%), Positives = 37/56 (65%)
Frame = +1
Query: 499 EFKVVDSETGEVLPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGW 554
E ++V +T + LPP G + +RGP +M+GY+ NP AT Q I+ GW TGD+G+
Sbjct: 31 ESQIVSLQTSKSLPPNQLGEIWLRGPVMMQGYFNNPEATKQTINDQGWTLTGDLGY 198
>TC89975 similar to PIR|G96530|G96530 probable acyl CoA synthetase
[imported] - Arabidopsis thaliana, partial (46%)
Length = 1251
Score = 59.7 bits (143), Expect = 3e-09
Identities = 46/132 (34%), Positives = 68/132 (50%), Gaps = 5/132 (3%)
Frame = +1
Query: 427 KLVYSKIHSAIGFS-KAGISGGGSLPSHVDRFFEA-IGVTLQNGYGLTET-SPVIAARRL 483
+LV+ K A+G + +S LP HV+ IG TL GYGLTE+ + A
Sbjct: 211 RLVFDKTKQALGGRVRILLSRAAPLPIHVEECLRVTIGSTLSQGYGLTESCAGCFTAIGD 390
Query: 484 SCNVIGSVGHPLKHTEFKVVD-SETG-EVLPPGYKGILKVRGPQLMKGYYKNPSATNQAI 541
+++G+VG P+ E ++ E G + L +G + +RG L GY+K T + I
Sbjct: 391 VFSMMGTVGIPMTTIEARLESVPEMGYDALSSEPRGEICLRGNSLFSGYHKRQDLTQEVI 570
Query: 542 DKDGWLNTGDIG 553
DGW +TGDIG
Sbjct: 571 -VDGWFHTGDIG 603
>TC89159 weakly similar to PIR|H85064|H85064 4-coumarate--CoA ligase-like
protein [imported] - Arabidopsis thaliana, partial (39%)
Length = 1103
Score = 57.8 bits (138), Expect = 1e-08
Identities = 52/198 (26%), Positives = 88/198 (44%), Gaps = 3/198 (1%)
Frame = +1
Query: 115 DKIALIDPYHDPPSTMTYKQLEDAILDFAEGLRVIGVSPNEKIALFADNSCRWLVADQGM 174
+K ALID D T+T+ QL+ + A L IG++ N+ + A NS + V +
Sbjct: 202 NKTALIDA--DSSQTLTFAQLKSLTIKLAHSLIKIGLTKNDVVLFLAPNSYLYPVFFLAV 375
Query: 175 MATGAI-NVVRGSRSSIEELLQIYNHSESIALAVDNPEMFNRIAKAFDLKASMRFVILLW 233
+ GA+ + V +++E Q + NP++ +A+ ++ +
Sbjct: 376 ASIGAVASTVNPQYTTVEVSKQAKD---------SNPKLIITVAELWEKVNHLNI----- 513
Query: 234 GEKSCLVNEGSKEVP--IFTFTEIMHLGRGSRRLFESHDARKHYVFEAIKSDDIATLVYT 291
+ +N P + +F + LG + F D +K D A L+Y+
Sbjct: 514 --PAVFLNSSGVATPDTVTSFESFVKLGESATE-FPKID---------VKQTDTAALLYS 657
Query: 292 SGTTGNPKGVMLTHQNLL 309
SGTTG KGV+LTHQN +
Sbjct: 658 SGTTGVSKGVILTHQNFI 711
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.326 0.140 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,581,081
Number of Sequences: 36976
Number of extensions: 293229
Number of successful extensions: 6894
Number of sequences better than 10.0: 143
Number of HSP's better than 10.0 without gapping: 3790
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5382
length of query: 617
length of database: 9,014,727
effective HSP length: 102
effective length of query: 515
effective length of database: 5,243,175
effective search space: 2700235125
effective search space used: 2700235125
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146706.7


