

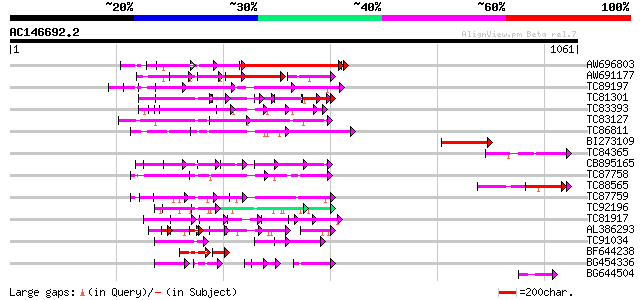
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146692.2 - phase: 0
(1061 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AW696803 similar to GP|14586365|e putative protein {Arabidopsis ... 410 e-116
AW691177 similar to GP|14586365|e putative protein {Arabidopsis ... 209 5e-54
TC89197 similar to GP|10177674|dbj|BAB11034. UVB-resistance prot... 179 7e-45
TC81301 similar to PIR|T50662|T50662 UVB-resistance protein UVR8... 120 3e-36
TC83393 similar to GP|15795108|dbj|BAB02372. gene_id:MJK13.9~unk... 142 9e-34
TC83127 similar to PIR|T47697|T47697 Regulator of chromosome con... 126 5e-29
TC86811 similar to GP|9279617|dbj|BAB01075.1 gb|AAF35409.1~gene_... 125 1e-28
BI273109 weakly similar to GP|9757959|db TMV resistance protein-... 113 4e-25
TC84365 similar to GP|15811367|gb|AAL08940.1 zinc finger protein... 112 8e-25
CB895165 similar to PIR|T47697|T47 Regulator of chromosome conde... 100 3e-21
TC87758 similar to PIR|T51387|T51387 UVB-resistance protein-like... 99 1e-20
TC88565 similar to GP|20197352|gb|AAC36161.2 expressed protein {... 96 1e-19
TC87759 similar to PIR|T51387|T51387 UVB-resistance protein-like... 93 5e-19
TC92196 similar to GP|16604485|gb|AAL24248.1 AT5g11580/F15N18_17... 86 8e-17
TC81917 similar to GP|10176914|dbj|BAB10107. contains similarity... 74 2e-13
AL386293 weakly similar to GP|4079809|gb| HERC2 {Homo sapiens}, ... 65 1e-10
TC91034 similar to GP|20161135|dbj|BAB90063. hypothetical protei... 62 2e-09
BF644238 similar to GP|10176914|dbj contains similarity to regul... 39 5e-08
BG454336 similar to GP|13548329|em putative protein {Arabidopsis... 44 1e-07
BG644504 similar to PIR|A86443|A864 probable major intrinsic pro... 53 7e-07
>AW696803 similar to GP|14586365|e putative protein {Arabidopsis thaliana},
partial (18%)
Length = 618
Score = 410 bits (1055), Expect(2) = e-116
Identities = 195/195 (100%), Positives = 195/195 (100%)
Frame = +3
Query: 430 TMRASCGVWHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCVALV 489
TMRASCGVWHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCVALV
Sbjct: 3 TMRASCGVWHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCVALV 182
Query: 490 EHNFCQVACGHSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEEIA 549
EHNFCQVACGHSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEEIA
Sbjct: 183 EHNFCQVACGHSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEEIA 362
Query: 550 CGAYHVAVLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAA 609
CGAYHVAVLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAA
Sbjct: 363 CGAYHVAVLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAA 542
Query: 610 ICLHKWVSGVDQSMC 624
ICLHKWVSGVDQSMC
Sbjct: 543 ICLHKWVSGVDQSMC 587
Score = 103 bits (256), Expect = 5e-22
Identities = 67/181 (37%), Positives = 91/181 (50%), Gaps = 12/181 (6%)
Frame = +3
Query: 275 ACGGRHAALVTK------------QGEIFSWGEESGGRLGHGVDSDVLHPKLIDALSNTN 322
+CG H A V + G++F+WG+ GRLGHG L P + AL N
Sbjct: 15 SCGVWHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCV-ALVEHN 191
Query: 323 IELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSYIS 382
VACG T A+T SG +Y G+ Y G LG+ Q +P RV G L V I+
Sbjct: 192 FCQVACGHSLTVALTTSGHVYAMGSPVY--GQLGNP-QADGKLPTRVEGKLLKSFVEEIA 362
Query: 383 CGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGVWHTAA 442
CG +H AV+T +++T+G G G LGHGD + P V++LK +CG TAA
Sbjct: 363 CGAYHVAVLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAA 542
Query: 443 V 443
+
Sbjct: 543 I 545
Score = 76.3 bits (186), Expect = 6e-14
Identities = 55/188 (29%), Positives = 85/188 (44%), Gaps = 3/188 (1%)
Frame = +3
Query: 207 SSAVSSSSQGSGHDDGDALGDVFIWGEGTGDGVVGGGNHRVGSGLGVKIDSLFPKALESA 266
++AV G+ + G +F WG+G G +G G+ K + +
Sbjct: 33 TAAVVEVMVGNSSSSNCSSGKLFTWGDGD-KGRLGHGDKEA-------------KLVPTC 170
Query: 267 VVLDVQN---IACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDALSNTNI 323
V L N +ACG +T G +++ G G+LG+ L ++ L + +
Sbjct: 171 VALVEHNFCQVACGHSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFV 350
Query: 324 ELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSYISC 383
E +ACG YH +TL ++YTWG GA G LGHG+ P V+ L+ HV I+C
Sbjct: 351 EEIACGAYHVAVLTLRNEVYTWGKGA--NGRLGHGDTDDRNNPTLVDA-LKDKHVKSIAC 521
Query: 384 GPWHTAVV 391
G TA +
Sbjct: 522 GTNFTAAI 545
Score = 70.9 bits (172), Expect = 3e-12
Identities = 34/94 (36%), Positives = 56/94 (59%), Gaps = 1/94 (1%)
Frame = +3
Query: 256 DSLFPKALESAVVLD-VQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKL 314
D P +E ++ V+ IACG H A++T + E+++WG+ + GRLGHG D +P L
Sbjct: 300 DGKLPTRVEGKLLKSFVEEIACGAYHVAVLTLRNEVYTWGKGANGRLGHGDTDDRNNPTL 479
Query: 315 IDALSNTNIELVACGEYHTCAVTLSGDLYTWGNG 348
+DAL + +++ +ACG T A+ L+ W +G
Sbjct: 480 VDALKDKHVKSIACGTNFTAAIC----LHKWVSG 569
Score = 28.1 bits (61), Expect(2) = e-116
Identities = 12/14 (85%), Positives = 13/14 (92%), Gaps = 1/14 (7%)
Frame = +1
Query: 622 SMC-SGCRLPFNFK 634
S+C SGCRLPFNFK
Sbjct: 577 SLCVSGCRLPFNFK 618
>AW691177 similar to GP|14586365|e putative protein {Arabidopsis thaliana},
partial (10%)
Length = 342
Score = 209 bits (532), Expect = 5e-54
Identities = 98/113 (86%), Positives = 108/113 (94%), Gaps = 1/113 (0%)
Frame = +2
Query: 404 TFGALGHGDRKSVSLPREVESLKGLRTMRASCGVWHTAAVVEVMVGNSSSSNCSSGKLFT 463
TFG LGHGDR+SVSLPRE+ESLKGLRT++A+CGVWHTAA+VEVMVGNSSSSNCSSGKLFT
Sbjct: 2 TFGVLGHGDRRSVSLPREIESLKGLRTVQAACGVWHTAAIVEVMVGNSSSSNCSSGKLFT 181
Query: 464 WGDGDKGRLGHGDKEAKLVPTC-VALVEHNFCQVACGHSLTVALTTSGHVYAM 515
WGDGDKGRLGHGDKE+KLVPTC VALVE NFCQVACGHS+TVAL+ +GHVY M
Sbjct: 182 WGDGDKGRLGHGDKESKLVPTCVVALVEPNFCQVACGHSMTVALSRAGHVYTM 340
Score = 67.0 bits (162), Expect = 4e-11
Identities = 37/104 (35%), Positives = 58/104 (55%), Gaps = 12/104 (11%)
Frame = +2
Query: 352 YGLLGHGNQVSHWVPKRVNGPLEGIHVSYISCGPWHTAVVT------------SAGQLFT 399
+G+LGHG++ S +P+ + L+G+ +CG WHTA + S+G+LFT
Sbjct: 5 FGVLGHGDRRSVSLPREIES-LKGLRTVQAACGVWHTAAIVEVMVGNSSSSNCSSGKLFT 181
Query: 400 FGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGVWHTAAV 443
+GDG G LGHGD++S +P V +L + +CG T A+
Sbjct: 182 WGDGDKGRLGHGDKESKLVPTCVVALVEPNFCQVACGHSMTVAL 313
Score = 58.5 bits (140), Expect = 1e-08
Identities = 39/119 (32%), Positives = 54/119 (44%), Gaps = 12/119 (10%)
Frame = +2
Query: 238 GVVGGGNHRVGSGLGVKIDSLFPKALESAVVLDVQNIACGGRHAALVTK----------- 286
GV+G G+ R S P+ +ES L ACG H A + +
Sbjct: 8 GVLGHGDRRSVS---------LPREIESLKGLRTVQAACGVWHTAAIVEVMVGNSSSSNC 160
Query: 287 -QGEIFSWGEESGGRLGHGVDSDVLHPKLIDALSNTNIELVACGEYHTCAVTLSGDLYT 344
G++F+WG+ GRLGHG L P + AL N VACG T A++ +G +YT
Sbjct: 161 SSGKLFTWGDGDKGRLGHGDKESKLVPTCVVALVEPNFCQVACGHSMTVALSRAGHVYT 337
Score = 57.4 bits (137), Expect = 3e-08
Identities = 38/113 (33%), Positives = 56/113 (48%), Gaps = 12/113 (10%)
Frame = +2
Query: 299 GRLGHGVDSDVLHPKLIDALSNTNIELVACGEYHTCAVT------------LSGDLYTWG 346
G LGHG V P+ I++L ACG +HT A+ SG L+TWG
Sbjct: 8 GVLGHGDRRSVSLPREIESLKGLRTVQAACGVWHTAAIVEVMVGNSSSSNCSSGKLFTWG 187
Query: 347 NGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSYISCGPWHTAVVTSAGQLFT 399
+G + G LGHG++ S VP V +E + ++CG T ++ AG ++T
Sbjct: 188 DG--DKGRLGHGDKESKLVPTCVVALVEP-NFCQVACGHSMTVALSRAGHVYT 337
Score = 53.9 bits (128), Expect = 3e-07
Identities = 34/105 (32%), Positives = 51/105 (48%), Gaps = 14/105 (13%)
Frame = +2
Query: 520 YGQLGN-PQADGKLPTRVEGKLLKSF-VEEIACGAYHVAVLT------------LRNEVY 565
+G LG+ + LP +E LK + ACG +H A + +++
Sbjct: 5 FGVLGHGDRRSVSLPREIES--LKGLRTVQAACGVWHTAAIVEVMVGNSSSSNCSSGKLF 178
Query: 566 TWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAAI 610
TWG G GRLGHGD + + PT V AL + + +ACG + T A+
Sbjct: 179 TWGDGDKGRLGHGDKESKLVPTCVVALVEPNFCQVACGHSMTVAL 313
Score = 37.7 bits (86), Expect = 0.024
Identities = 33/111 (29%), Positives = 50/111 (44%), Gaps = 14/111 (12%)
Frame = +2
Query: 470 GRLGHGDKEAKLVPTCV-ALVEHNFCQVACGHSLTVALT------------TSGHVYAMG 516
G LGHGD+ + +P + +L Q ACG T A+ +SG ++ G
Sbjct: 8 GVLGHGDRRSVSLPREIESLKGLRTVQAACGVWHTAAIVEVMVGNSSSSNCSSGKLFTWG 187
Query: 517 SPVYGQLGNPQADGKL-PTRVEGKLLKSFVEEIACGAYHVAVLTLRNEVYT 566
G+LG+ + KL PT V + +F ++ACG L+ VYT
Sbjct: 188 DGDKGRLGHGDKESKLVPTCVVALVEPNFC-QVACGHSMTVALSRAGHVYT 337
>TC89197 similar to GP|10177674|dbj|BAB11034. UVB-resistance protein UVR8
{Arabidopsis thaliana}, partial (74%)
Length = 1457
Score = 179 bits (453), Expect = 7e-45
Identities = 123/362 (33%), Positives = 182/362 (49%), Gaps = 9/362 (2%)
Frame = +3
Query: 274 IACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDALSNTNIELVACGEYHT 333
I+ G H + + SWG G+LGHG D L P + A +I V CG +T
Sbjct: 60 ISAGASHTVALLTGNVVCSWGRGEDGQLGHGDTDDRLLPTKLSAFDGQDIVSVTCGADYT 239
Query: 334 CAVTLSG-DLYTWGN-GAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSYISCGPWHTAVV 391
A + SG D+Y+WG G G LG G VP+++ EGI + ++ G H+ +
Sbjct: 240 VARSKSGKDVYSWGCWGRNQNGELGLGTTKDSHVPQKILA-FEGIRIKMVAAGAEHSVAI 416
Query: 392 TSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGVWHTAAVVEVMVGNS 451
T G L+ +G G +G LG GD +P +V ++ G + + SCG HT ++
Sbjct: 417 TEDGDLYGWGWGRYGNLGLGDTNDRLIPEKV-NIDGDKIVMVSCGWRHTISI-------- 569
Query: 452 SSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCV-ALVEHNFCQVACGHSLTVALTTSG 510
SSG L+T G G+LGHGD E LVP V AL + QV+ G ++ALT++G
Sbjct: 570 ----SSSGGLYTHGWSKYGQLGHGDFEDCLVPRKVQALSDKLISQVSGGWRHSMALTSNG 737
Query: 511 HVYAMGSPVYGQLG-NPQADGKLPTRVEGKLLKSFVEEIACGAYHVAVLTLRNEVYTWGK 569
++ G +GQ+G D P V + V +++ G H +T R VY+WG+
Sbjct: 738 QLFGWGWNKFGQIGIGHNFDCSSPMLVNFPHAQKVV-QMSSGWRHTVAVTDRANVYSWGR 914
Query: 570 GANGRLGHGDTDDRNNPTLVDAL-----KDKHVKSIACGTNFTAAICLHKWVSGVDQSMC 624
GANG+LGHGDT DRN PT++DA ++H++S + L + +G + C
Sbjct: 915 GANGQLGHGDTKDRNVPTIIDAFSVDGCSEQHIESSNYPLPGKSFASLSERYAGCSK*NC 1094
Query: 625 SG 626
SG
Sbjct: 1095SG 1100
Score = 143 bits (361), Expect = 3e-34
Identities = 106/331 (32%), Positives = 158/331 (47%), Gaps = 7/331 (2%)
Frame = +3
Query: 214 SQGSGHDDGDALGDVFI-WGEGTGDGVVGGGNHRVGSGLGVKIDSLFPKALESAVVLDVQ 272
S G+ H G+V WG G DG +G G+ D L P L + D+
Sbjct: 63 SAGASHTVALLTGNVVCSWGRGE-DGQLGHGDTD---------DRLLPTKLSAFDGQDIV 212
Query: 273 NIACGGRHAALVTKQG-EIFSWG---EESGGRLGHGVDSDVLHPKLIDALSNTNIELVAC 328
++ CG + +K G +++SWG G LG G D P+ I A I++VA
Sbjct: 213 SVTCGADYTVARSKSGKDVYSWGCWGRNQNGELGLGTTKDSHVPQKILAFEGIRIKMVAA 392
Query: 329 GEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSYISCGPWHT 388
G H+ A+T GDLY WG G YG LG G+ +P++VN ++G + +SCG HT
Sbjct: 393 GAEHSVAITEDGDLYGWGWG--RYGNLGLGDTNDRLIPEKVN--IDGDKIVMVSCGWRHT 560
Query: 389 AVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGVWHTAAVVEVMV 448
++S+G L+T G +G LGHGD + +PR+V++L + S G H+ A+
Sbjct: 561 ISISSSGGLYTHGWSKYGQLGHGDFEDCLVPRKVQALSDKLISQVSGGWRHSMALT---- 728
Query: 449 GNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCVALVE-HNFCQVACGHSLTVALT 507
S+G+LF WG G++G G P V Q++ G TVA+T
Sbjct: 729 --------SNGQLFGWGWNKFGQIGIGHNFDCSSPMLVNFPHAQKVVQMSSGWRHTVAVT 884
Query: 508 TSGHVYAMGSPVYGQLGNPQA-DGKLPTRVE 537
+VY+ G GQLG+ D +PT ++
Sbjct: 885 DRANVYSWGRGANGQLGHGDTKDRNVPTIID 977
Score = 99.0 bits (245), Expect = 9e-21
Identities = 72/244 (29%), Positives = 115/244 (46%), Gaps = 4/244 (1%)
Frame = +3
Query: 185 GGSDSMHGHMKTMGMDAFRVSLSSAVSSSSQGSGHDDGDALGDVFIWGEGTGDGVVGGGN 244
G + H K + + R+ + +A + S D GD++ WG G
Sbjct: 318 GTTKDSHVPQKILAFEGIRIKMVAAGAEHSVAITED-----GDLYGWGWG---------- 452
Query: 245 HRVGS-GLGVKIDSLFPKALE---SAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGR 300
R G+ GLG D L P+ + +V+ ++CG RH ++ G +++ G G+
Sbjct: 453 -RYGNLGLGDTNDRLIPEKVNIDGDKIVM----VSCGWRHTISISSSGGLYTHGWSKYGQ 617
Query: 301 LGHGVDSDVLHPKLIDALSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQ 360
LGHG D L P+ + ALS+ I V+ G H+ A+T +G L+ WG +G +G G+
Sbjct: 618 LGHGDFEDCLVPRKVQALSDKLISQVSGGWRHSMALTSNGQLFGWGWN--KFGQIGIGHN 791
Query: 361 VSHWVPKRVNGPLEGIHVSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPR 420
P VN P V +S G HT VT ++++G G G LGHGD K ++P
Sbjct: 792 FDCSSPMLVNFP-HAQKVVQMSSGWRHTVAVTDRANVYSWGRGANGQLGHGDTKDRNVPT 968
Query: 421 EVES 424
+++
Sbjct: 969 IIDA 980
Score = 50.1 bits (118), Expect = 5e-06
Identities = 30/93 (32%), Positives = 43/93 (45%), Gaps = 1/93 (1%)
Frame = +3
Query: 241 GGGNHRVGS-GLGVKIDSLFPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGG 299
G G ++ G G+G D P + V ++ G RH VT + ++SWG + G
Sbjct: 747 GWGWNKFGQIGIGHNFDCSSPMLVNFPHAQKVVQMSSGWRHTVAVTDRANVYSWGRGANG 926
Query: 300 RLGHGVDSDVLHPKLIDALSNTNIELVACGEYH 332
+LGHG D P +IDA S + C E H
Sbjct: 927 QLGHGDTKDRNVPTIIDAFS-----VDGCSEQH 1010
>TC81301 similar to PIR|T50662|T50662 UVB-resistance protein UVR8 [imported]
- Arabidopsis thaliana, partial (70%)
Length = 1253
Score = 120 bits (301), Expect = 3e-27
Identities = 68/203 (33%), Positives = 108/203 (52%)
Frame = +1
Query: 274 IACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDALSNTNIELVACGEYHT 333
+ G H VT +GE+ SWG G+LG G D L P+ I I++VA G H+
Sbjct: 637 LLAGDSHCLAVTMEGEVQSWGRNQNGQLGLGTTEDSLVPQKIQTFQGVPIKMVAAGAEHS 816
Query: 334 CAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSYISCGPWHTAVVTS 393
A+T +G+LY WG G YG LG G++ +P++V+ ++ + ++CG HT V+S
Sbjct: 817 VAITENGELYGWGWG--RYGNLGLGDRNDRCIPEKVS--VDCDKMVLVACGWRHTISVSS 984
Query: 394 AGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGVWHTAAVVEVMVGNSSS 453
+G+L+T+G +G LGHGD K +P ++++ + S G HT A+
Sbjct: 985 SGELYTYGWSKYGQLGHGDCKDHLVPHKLQAFSDKLICQVSGGWRHTMALT--------- 1137
Query: 454 SNCSSGKLFTWGDGDKGRLGHGD 476
+ G+L+ WG G++G GD
Sbjct: 1138---TGGQLYGWGWNKFGQVGVGD 1197
Score = 103 bits (256), Expect = 5e-22
Identities = 60/171 (35%), Positives = 95/171 (55%)
Frame = +1
Query: 242 GGNHRVGSGLGVKIDSLFPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRL 301
G N GLG DSL P+ +++ + ++ +A G H+ +T+ GE++ WG G L
Sbjct: 697 GRNQNGQLGLGTTEDSLVPQKIQTFQGVPIKMVAAGAEHSVAITENGELYGWGWGRYGNL 876
Query: 302 GHGVDSDVLHPKLIDALSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQV 361
G G +D P+ + ++ + LVACG HT +V+ SG+LYT+G YG LGHG+
Sbjct: 877 GLGDRNDRCIPEKV-SVDCDKMVLVACGWRHTISVSSSGELYTYGWS--KYGQLGHGDCK 1047
Query: 362 SHWVPKRVNGPLEGIHVSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGD 412
H VP ++ + + + +S G HT +T+ GQL+ +G FG +G GD
Sbjct: 1048DHLVPHKLQAFSDKL-ICQVSGGWRHTMALTTGGQLYGWGWNKFGQVGVGD 1197
Score = 97.4 bits (241), Expect(2) = 3e-36
Identities = 60/209 (28%), Positives = 102/209 (48%), Gaps = 4/209 (1%)
Frame = +1
Query: 379 SYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGVW 438
S + G H VT G++ ++G G LG G + +P+++++ +G+ + G
Sbjct: 631 SKLLAGDSHCLAVTMEGEVQSWGRNQNGQLGLGTTEDSLVPQKIQTFQGVPIKMVAAGAE 810
Query: 439 HTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCVALVEHNFCQVAC 498
H+ A+ E +G+L+ WG G G LG GD+ + +P V++ VAC
Sbjct: 811 HSVAITE------------NGELYGWGWGRYGNLGLGDRNDRCIPEKVSVDCDKMVLVAC 954
Query: 499 GHSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEEIAC----GAYH 554
G T+++++SG +Y G YGQLG+ G + L++F +++ C G H
Sbjct: 955 GWRHTISVSSSGELYTYGWSKYGQLGH----GDCKDHLVPHKLQAFSDKLICQVSGGWRH 1122
Query: 555 VAVLTLRNEVYTWGKGANGRLGHGDTDDR 583
LT ++Y WG G++G GD DR
Sbjct: 1123TMALTTGGQLYGWGWNKFGQVGVGDNVDR 1209
Score = 75.5 bits (184), Expect = 1e-13
Identities = 50/154 (32%), Positives = 75/154 (48%), Gaps = 2/154 (1%)
Frame = +1
Query: 459 GKLFTWGDGDKGRLGHGDKEAKLVPTCVALVEH-NFCQVACGHSLTVALTTSGHVYAMGS 517
G++ +WG G+LG G E LVP + + VA G +VA+T +G +Y G
Sbjct: 679 GEVQSWGRNQNGQLGLGTTEDSLVPQKIQTFQGVPIKMVAAGAEHSVAITENGELYGWGW 858
Query: 518 PVYGQLG-NPQADGKLPTRVEGKLLKSFVEEIACGAYHVAVLTLRNEVYTWGKGANGRLG 576
YG LG + D +P +V K + +ACG H ++ E+YT+G G+LG
Sbjct: 859 GRYGNLGLGDRNDRCIPEKVSVDCDKMVL--VACGWRHTISVSSSGELYTYGWSKYGQLG 1032
Query: 577 HGDTDDRNNPTLVDALKDKHVKSIACGTNFTAAI 610
HGD D P + A DK + ++ G T A+
Sbjct: 1033HGDCKDHLVPHKLQAFSDKLICQVSGGWRHTMAL 1134
Score = 73.9 bits (180), Expect(2) = 3e-36
Identities = 40/111 (36%), Positives = 60/111 (54%), Gaps = 1/111 (0%)
Frame = +3
Query: 274 IACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDALSNTNIELVACGEYHT 333
I+ GG H+ + + SWG G+LGHG D L P + AL I V CG HT
Sbjct: 321 ISAGGSHSVALISGNVVCSWGRGEDGQLGHGDTDDRLLPTQLSALDAQQIVSVVCGADHT 500
Query: 334 CAVTLSG-DLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSYISC 383
A + S ++Y+WG G ++G LGHGN ++P+ + L+G+ + I+C
Sbjct: 501 LAYSESKVEVYSWGWG--DFGRLGHGNSSDLFIPQPIKA-LQGLRIKQIAC 644
Score = 63.9 bits (154), Expect = 3e-10
Identities = 41/109 (37%), Positives = 60/109 (54%), Gaps = 2/109 (1%)
Frame = +3
Query: 496 VACGHSLTVALTTSGHVYAMGSPVYGQLGNPQADGKL-PTRVEGKLLKSFVEEIACGAYH 554
++ G S +VAL + V + G GQLG+ D +L PT++ + V + CGA H
Sbjct: 321 ISAGGSHSVALISGNVVCSWGRGEDGQLGHGDTDDRLLPTQLSALDAQQIVS-VVCGADH 497
Query: 555 -VAVLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIAC 602
+A + EVY+WG G GRLGHG++ D P + AL+ +K IAC
Sbjct: 498 TLAYSESKVEVYSWGWGDFGRLGHGNSSDLFIPQPIKALQGLRIKQIAC 644
Score = 63.5 bits (153), Expect = 4e-10
Identities = 40/122 (32%), Positives = 58/122 (46%), Gaps = 1/122 (0%)
Frame = +3
Query: 378 VSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGV 437
V IS G H+ + S + ++G G G LGHGD LP ++ +L + + CG
Sbjct: 312 VLLISAGGSHSVALISGNVVCSWGRGEDGQLGHGDTDDRLLPTQLSALDAQQIVSVVCGA 491
Query: 438 WHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCV-ALVEHNFCQV 496
HT A E S ++++WG GD GRLGHG+ +P + AL Q+
Sbjct: 492 DHTLAYSE-----------SKVEVYSWGWGDFGRLGHGNSSDLFIPQPIKALQGLRIKQI 638
Query: 497 AC 498
AC
Sbjct: 639 AC 644
Score = 61.2 bits (147), Expect = 2e-09
Identities = 28/62 (45%), Positives = 38/62 (61%)
Frame = +3
Query: 548 IACGAYHVAVLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFT 607
I+ G H L N V +WG+G +G+LGHGDTDDR PT + AL + + S+ CG + T
Sbjct: 321 ISAGGSHSVALISGNVVCSWGRGEDGQLGHGDTDDRLLPTQLSALDAQQIVSVVCGADHT 500
Query: 608 AA 609
A
Sbjct: 501 LA 506
Score = 45.8 bits (107), Expect = 9e-05
Identities = 20/64 (31%), Positives = 33/64 (51%)
Frame = +1
Query: 547 EIACGAYHVAVLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNF 606
++ G H +T+ EV +WG+ NG+LG G T+D P + + +K +A G
Sbjct: 634 KLLAGDSHCLAVTMEGEVQSWGRNQNGQLGLGTTEDSLVPQKIQTFQGVPIKMVAAGAEH 813
Query: 607 TAAI 610
+ AI
Sbjct: 814 SVAI 825
Score = 31.2 bits (69), Expect = 2.2
Identities = 15/53 (28%), Positives = 27/53 (50%)
Frame = +1
Query: 256 DSLFPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSD 308
D L P L++ + ++ G RH +T G+++ WG G++G G + D
Sbjct: 1048 DHLVPHKLQAFSDKLICQVSGGWRHTMALTTGGQLYGWGWNKFGQVGVGDNVD 1206
>TC83393 similar to GP|15795108|dbj|BAB02372. gene_id:MJK13.9~unknown
protein {Arabidopsis thaliana}, partial (59%)
Length = 1018
Score = 142 bits (357), Expect = 9e-34
Identities = 95/306 (31%), Positives = 147/306 (47%), Gaps = 8/306 (2%)
Frame = +3
Query: 280 HAALVTKQGEIFSWGEESGGRLGH-GVDSDVLHPKLIDALSNTNIELVACGEYHTCAVTL 338
HAA V + GE+F+ G+ S GH + + P+L+++L + V+ G T +T
Sbjct: 12 HAAFVMESGEVFTCGDNSSFCCGHKDTNRPIFRPRLVESLKGIPCKQVSAGLNFTVFLTR 191
Query: 339 SGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSYISCGPWHTAVVTSAGQLF 398
G +YT G+ ++ G LGH + PK++ + ++ GP + VT G ++
Sbjct: 192 QGHVYTCGSNSH--GQLGHSDTQDRPSPKKIEVLGSIGRIVQVAAGPNYILSVTEGGSVY 365
Query: 399 TFGDGTFGALGHGDRKSVSLPREVESLK--GLRTMRASCGVWHTAAVVEVMVGNSSSSNC 456
+FG G+ LGHG++ LPR ++ + G+ + S G H AA+
Sbjct: 366 SFGSGSNFCLGHGEQHDELLPRCIQKFRRNGIHVVHVSAGDEHAAALD------------ 509
Query: 457 SSGKLFTWGDGDKGRLGHGDKEAKLVPTCVA-LVEHNFCQVACGHSLTVALTTSGHVYAM 515
S+G ++TWG G G LGHGD+ K P ++ L H QV T L SG VY
Sbjct: 510 SNGFVYTWGKGYCGALGHGDEIEKTRPELLSSLKNHLAVQVCAKKRKTFVLVDSGLVYGF 689
Query: 516 GSPVYGQLGNPQA----DGKLPTRVEGKLLKSFVEEIACGAYHVAVLTLRNEVYTWGKGA 571
GS +G LG P A + L R+ L V +I+ G YH V+T R ++ +G
Sbjct: 690 GSMAFGSLGFPDARRLTNKILKPRIVYTLRTHHVSQISTGLYHTVVVTNRGKILGFGDNE 869
Query: 572 NGRLGH 577
+LGH
Sbjct: 870 RAQLGH 887
Score = 110 bits (275), Expect = 3e-24
Identities = 83/273 (30%), Positives = 129/273 (46%), Gaps = 7/273 (2%)
Frame = +3
Query: 260 PKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDALS 319
P+ +ES + + ++ G +T+QG +++ G S G+LGH D PK I+ L
Sbjct: 111 PRLVESLKGIPCKQVSAGLNFTVFLTRQGHVYTCGSNSHGQLGHSDTQDRPSPKKIEVLG 290
Query: 320 NTN-IELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNG-PLEGIH 377
+ I VA G + +VT G +Y++G+G+ N+ LGHG Q +P+ + GIH
Sbjct: 291 SIGRIVQVAAGPNYILSVTEGGSVYSFGSGS-NF-CLGHGEQHDELLPRCIQKFRRNGIH 464
Query: 378 VSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGV 437
V ++S G H A + S G ++T+G G GALGHGD + P + SLK ++
Sbjct: 465 VVHVSAGDEHAAALDSNGFVYTWGKGYCGALGHGDEIEKTRPELLSSLKNHLAVQVCAKK 644
Query: 438 WHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAK----LVPTCV-ALVEHN 492
T +V+ SG ++ +G G LG D L P V L H+
Sbjct: 645 RKTFVLVD------------SGLVYGFGSMAFGSLGFPDARRLTNKILKPRIVYTLRTHH 788
Query: 493 FCQVACGHSLTVALTTSGHVYAMGSPVYGQLGN 525
Q++ G TV +T G + G QLG+
Sbjct: 789 VSQISTGLYHTVVVTNRGKILGFGDNERAQLGH 887
Score = 102 bits (255), Expect = 6e-22
Identities = 68/213 (31%), Positives = 103/213 (47%), Gaps = 5/213 (2%)
Frame = +3
Query: 387 HTAVVTSAGQLFTFGDGTFGALGHGD-RKSVSLPREVESLKGLRTMRASCGVWHTAAVVE 445
H A V +G++FT GD + GH D + + PR VESLKG+ + S G+ T +
Sbjct: 12 HAAFVMESGEVFTCGDNSSFCCGHKDTNRPIFRPRLVESLKGIPCKQVSAGLNFTVFLTR 191
Query: 446 VMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCVALVEH--NFCQVACGHSLT 503
G ++T G G+LGH D + + P + ++ QVA G +
Sbjct: 192 ------------QGHVYTCGSNSHGQLGHSDTQDRPSPKKIEVLGSIGRIVQVAAGPNYI 335
Query: 504 VALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKS--FVEEIACGAYHVAVLTLR 561
+++T G VY+ GS LG+ + +L R K ++ V ++ G H A L
Sbjct: 336 LSVTEGGSVYSFGSGSNFCLGHGEQHDELLPRCIQKFRRNGIHVVHVSAGDEHAAALDSN 515
Query: 562 NEVYTWGKGANGRLGHGDTDDRNNPTLVDALKD 594
VYTWGKG G LGHGD ++ P L+ +LK+
Sbjct: 516 GFVYTWGKGYCGALGHGDEIEKTRPELLSSLKN 614
Score = 93.6 bits (231), Expect = 4e-19
Identities = 63/220 (28%), Positives = 104/220 (46%), Gaps = 6/220 (2%)
Frame = +3
Query: 271 VQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDALSNTNIELV--AC 328
+ +A G + VT+ G ++S+G S LGHG D L P+ I I +V +
Sbjct: 303 IVQVAAGPNYILSVTEGGSVYSFGSGSNFCLGHGEQHDELLPRCIQKFRRNGIHVVHVSA 482
Query: 329 GEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSYISCGPWHT 388
G+ H A+ +G +YTWG G G LGHG+++ P+ ++ + V + T
Sbjct: 483 GDEHAAALDSNGFVYTWGKGYC--GALGHGDEIEKTRPELLSSLKNHLAVQ-VCAKKRKT 653
Query: 389 AVVTSAGQLFTFGDGTFGALGHGDRKSVS----LPREVESLKGLRTMRASCGVWHTAAVV 444
V+ +G ++ FG FG+LG D + ++ PR V +L+ + S G++HT V
Sbjct: 654 FVLVDSGLVYGFGSMAFGSLGFPDARRLTNKILKPRIVYTLRTHHVSQISTGLYHTVVVT 833
Query: 445 EVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPT 484
+ GK+ +GD ++ +LGH L PT
Sbjct: 834 ------------NRGKILGFGDNERAQLGHDTLRHCLEPT 917
Score = 82.0 bits (201), Expect = 1e-15
Identities = 56/191 (29%), Positives = 93/191 (48%), Gaps = 9/191 (4%)
Frame = +3
Query: 241 GGGNHRVGSGL------GVKIDSLFPKALES--AVVLDVQNIACGGRHAALVTKQGEIFS 292
GG + GSG G + D L P+ ++ + V +++ G HAA + G +++
Sbjct: 351 GGSVYSFGSGSNFCLGHGEQHDELLPRCIQKFRRNGIHVVHVSAGDEHAAALDSNGFVYT 530
Query: 293 WGEESGGRLGHGVDSDVLHPKLIDALSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNY 352
WG+ G LGHG + + P+L+ +L N V + T + SG +Y +G+ A+
Sbjct: 531 WGKGYCGALGHGDEIEKTRPELLSSLKNHLAVQVCAKKRKTFVLVDSGLVYGFGSMAFGS 710
Query: 353 GLLGHGNQVSHWVPK-RVNGPLEGIHVSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHG 411
++++ + K R+ L HVS IS G +HT VVT+ G++ FGD LGH
Sbjct: 711 LGFPDARRLTNKILKPRIVYTLRTHHVSQISTGLYHTVVVTNRGKILGFGDNERAQLGHD 890
Query: 412 DRKSVSLPREV 422
+ P E+
Sbjct: 891 TLRHCLEPTEI 923
>TC83127 similar to PIR|T47697|T47697 Regulator of chromosome
condensation-like protein - Arabidopsis thaliana,
partial (46%)
Length = 1208
Score = 126 bits (316), Expect = 5e-29
Identities = 91/272 (33%), Positives = 132/272 (48%), Gaps = 23/272 (8%)
Frame = +3
Query: 204 VSLSSAVSSSSQGSGHDDGDALGDVF-IWGEGTGDGVVGGGNHRVGSGLGVKIDSL--FP 260
V+L V +S G AL DV +WG G G G ++G G VK+ S
Sbjct: 129 VTLGHGVKITSVAVGGRHTLALSDVGQVWGWGYG------GEGQLGLGSRVKMVSSPHLI 290
Query: 261 KALESAVVLD--------------------VQNIACGGRHAALVTKQGEIFSWGEESGGR 300
+ESA D V IACGGRH+A++T G + ++G G+
Sbjct: 291 SCIESASGKDKSSFQQGSTALQCSNVAGNYVTEIACGGRHSAVITDTGALLTFGWGLHGQ 470
Query: 301 LGHGVDSDVLHPKLIDALSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQ 360
G G ++D L P L+ +L T ++ +A G +HT VT +G LY +G +G LG GN
Sbjct: 471 CGQGNNADQLRPTLVPSLLGTRVKQIAAGLWHTLCVTANGQLYAFGGN--QFGQLGTGND 644
Query: 361 VSHWVPKRVNGPLEGIHVSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPR 420
+ PK ++ E S +SCG H+A++T G LFT+G +G LG GD ++P
Sbjct: 645 QTETSPKLLD-TFENNLSSIVSCGARHSALLTDDGHLFTWGWNKYGQLGLGDSVDRNIPS 821
Query: 421 EVESLKGLRTMRASCGVWHTAAVVEVMVGNSS 452
V S+ G R +CG WHT + + V +S
Sbjct: 822 RV-SISGCRPRNVACGWWHTLLMADKPV*RTS 914
Score = 103 bits (256), Expect = 5e-22
Identities = 78/246 (31%), Positives = 122/246 (48%), Gaps = 17/246 (6%)
Frame = +3
Query: 375 GIHVSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDR-KSVSLPREVESLKGLRTMRA 433
G+ ++ ++ G HT ++ GQ++ +G G G LG G R K VS P + ++
Sbjct: 144 GVKITSVAVGGRHTLALSDVGQVWGWGYGGEGQLGLGSRVKMVSSPHLISCIESASGKDK 323
Query: 434 SCGVWHTAA----------VVEVMVGNSSSSNCS-SGKLFTWGDGDKGRLGHGDKEAKLV 482
S + A V E+ G S+ + +G L T+G G G+ G G+ +L
Sbjct: 324 SSFQQGSTALQCSNVAGNYVTEIACGGRHSAVITDTGALLTFGWGLHGQCGQGNNADQLR 503
Query: 483 PTCV-ALVEHNFCQVACGHSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLL 541
PT V +L+ Q+A G T+ +T +G +YA G +GQLG G T KLL
Sbjct: 504 PTLVPSLLGTRVKQIAAGLWHTLCVTANGQLYAFGGNQFGQLGT----GNDQTETSPKLL 671
Query: 542 KSFVEE----IACGAYHVAVLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHV 597
+F ++CGA H A+LT ++TWG G+LG GD+ DRN P+ V ++
Sbjct: 672 DTFENNLSSIVSCGARHSALLTDDGHLFTWGWNKYGQLGLGDSVDRNIPSRV-SISGCRP 848
Query: 598 KSIACG 603
+++ACG
Sbjct: 849 RNVACG 866
>TC86811 similar to GP|9279617|dbj|BAB01075.1
gb|AAF35409.1~gene_id:MPE11.29~similar to unknown protein
{Arabidopsis thaliana}, partial (94%)
Length = 2156
Score = 125 bits (313), Expect = 1e-28
Identities = 102/321 (31%), Positives = 157/321 (48%), Gaps = 12/321 (3%)
Frame = +1
Query: 338 LSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSYISCGPWHTAVVTSAGQL 397
L GD+Y++G+ + G LGHG W PK + L+GI + + G T ++T +GQ+
Sbjct: 727 LKGDVYSFGSNSS--GQLGHGTTEEEWRPKPIR-TLQGIRIIQAAAGAGRTMLITDSGQV 897
Query: 398 FTFGDGTFGALGHGDR--KSVSLPREVESLKGLRTMRASCGVWHTAAVVEVMVGNSSSSN 455
+ FG +FG +G + K+V PR VESLK + ++A+ G + TA +
Sbjct: 898 YAFGKDSFGEAEYGGQGSKTVIAPRLVESLKNIFVVQAAIGNFFTAVLSR---------- 1047
Query: 456 CSSGKLFTWGDGDKGRLGH--GDKEAKLVPTCVALVEHNFCQVACGHS--LTVALTTSG- 510
G+++T+ G G+LGH + + P AL + Q+A G+ L +A SG
Sbjct: 1048 --EGRVYTFSWGSDGKLGHHTDSNDMEPHPLLGALEDIPVVQIAAGYCYLLCLACQPSGM 1221
Query: 511 HVYAMGSPVYGQLGN-PQADGKLPTRVEG-KLLKSFVEEIACGAYHVAVLTLRNEVYTWG 568
VY++G + G+LG+ + D K P +E + L I+ GA+H AV+ V TWG
Sbjct: 1222 SVYSVGCGLGGKLGHGTRTDEKFPRLIEQFQTLNLQPMVISAGAWHAAVVGRDGRVCTWG 1401
Query: 569 KGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACG--TNFTAAICLHKWVSGVDQSMCSG 626
G G LGHG+ + P +VDAL + +A G T F A + G +S G
Sbjct: 1402 WGRYGCLGHGNEECEAAPKVVDALTNVKAVHVATGDYTTFVVAEDGDVYSFGCGESASLG 1581
Query: 627 CRLPFNFKRKRH-NCYNCGLV 646
+ + RH N N LV
Sbjct: 1582 HNAVIDAEGNRHANVLNPELV 1644
Score = 110 bits (276), Expect = 2e-24
Identities = 88/321 (27%), Positives = 149/321 (46%), Gaps = 22/321 (6%)
Frame = +1
Query: 226 GDVFIWGEGT-GDGVVGGGNHRVGSGLGVKIDSLFPKALESAVVLDVQNIACGGRHAALV 284
G V+ +G+ + G+ GG + + P+ +ES + V A G A++
Sbjct: 889 GQVYAFGKDSFGEAEYGGQGSKT---------VIAPRLVESLKNIFVVQAAIGNFFTAVL 1041
Query: 285 TKQGEI--FSWGEESGGRLGHGVDSDVLHPK-LIDALSNTNIELVACGE-YHTCAVTLSG 340
+++G + FSWG S G+LGH DS+ + P L+ AL + + +A G Y C
Sbjct: 1042 SREGRVYTFSWG--SDGKLGHHTDSNDMEPHPLLGALEDIPVVQIAAGYCYLLCLACQPS 1215
Query: 341 DLYTWGNGAYNYGLLGHGNQVSHWVPKRVNG-PLEGIHVSYISCGPWHTAVVTSAGQLFT 399
+ + G G LGHG + P+ + + IS G WH AVV G++ T
Sbjct: 1216 GMSVYSVGCGLGGKLGHGTRTDEKFPRLIEQFQTLNLQPMVISAGAWHAAVVGRDGRVCT 1395
Query: 400 FGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGVWHTAAVVEVMVGNSSSSNCSSG 459
+G G +G LGHG+ + + P+ V++L ++ + + G + T V E G
Sbjct: 1396 WGWGRYGCLGHGNEECEAAPKVVDALTNVKAVHVATGDYTTFVVAE------------DG 1539
Query: 460 KLFTWGDGDKGRLGH-------GDKEAKL----VPTCVALVEHNFCQVACGHSL-----T 503
++++G G+ LGH G++ A + + T + + Q++ +S+ T
Sbjct: 1540 DVYSFGCGESASLGHNAVIDAEGNRHANVLNPELVTSLKQINERVVQISLTNSIYWNAHT 1719
Query: 504 VALTTSGHVYAMGSPVYGQLG 524
ALT SG +YA G+ GQLG
Sbjct: 1720 FALTESGKLYAFGAGDKGQLG 1782
>BI273109 weakly similar to GP|9757959|db TMV resistance protein-like
{Arabidopsis thaliana}, partial (10%)
Length = 589
Score = 113 bits (282), Expect = 4e-25
Identities = 55/94 (58%), Positives = 76/94 (80%)
Frame = +3
Query: 809 TPTPTLGGLTTPKIVVDDAKKTNDSLSQEVIKLRSQVESLTRKAQLQEIELERTSKQLKD 868
TP PT+ GL+ K + D KKTN+ L++EV KL +QVESL K + QE+E++R++K+ ++
Sbjct: 3 TPIPTMSGLSFSKNIADSLKKTNELLNKEVQKLHAQVESLKNKCERQELEVQRSAKKTQE 182
Query: 869 AIAIAGEETAKCKAAKEVIKSLTAQLKDMAERLP 902
AIA+A EE+ KCKAAK+VIKSLTAQLKD+AE+LP
Sbjct: 183 AIALATEESTKCKAAKQVIKSLTAQLKDLAEKLP 284
>TC84365 similar to GP|15811367|gb|AAL08940.1 zinc finger protein {Arabidopsis
thaliana}, partial (6%)
Length = 597
Score = 112 bits (280), Expect = 8e-25
Identities = 73/168 (43%), Positives = 95/168 (56%), Gaps = 6/168 (3%)
Frame = +2
Query: 890 LTAQLKDMAERLPVGT-AKSVKSPSIASFGSNELSFAAIDRLN-----IQATSPEADLTG 943
L + L + ER G+ A ++ S AS SN DRL IQAT+
Sbjct: 8 LESSLMNRTERNSTGSYATNLYQQSRASVTSNGTDDYRDDRLPNSGSMIQATN------- 166
Query: 944 SNTQLLSNGSSTVSNRSTGQNKQSQSDSTNRNGSRTKDSESRSETEWVEQDEPGVYITLT 1003
S+ S T R++G + +S S +RN +S ++ E EW+EQ EPGVYITL
Sbjct: 167 ------SSVSDTFDGRNSGNFRDDESGSRSRNDVLAANS-NQVEAEWIEQYEPGVYITLV 325
Query: 1004 SLPGGVIDLKRVRFSRKRFSEKQAENWWAENRVRVYEQYNVRMVDKSS 1051
++ G DL+RVRFSR+RF E QAE WW+ENR RVYE+YNVR DKSS
Sbjct: 326 AMRDGTRDLRRVRFSRRRFGENQAETWWSENRDRVYERYNVRSSDKSS 469
>CB895165 similar to PIR|T47697|T47 Regulator of chromosome condensation-like
protein - Arabidopsis thaliana, partial (31%)
Length = 853
Score = 100 bits (249), Expect = 3e-21
Identities = 56/159 (35%), Positives = 88/159 (55%), Gaps = 1/159 (0%)
Frame = +2
Query: 288 GEIFSWGEESGGRLGHGVDSDVLHPKLIDALSNTNIELVACGEYHTCAVTLSGDLYTWGN 347
G + ++G G+ G G D L P + +L IE VA G +HT + GD+Y +G
Sbjct: 5 GAVLAFGWGLYGQCGQGSTDDELSPTCVSSLLGIQIEGVAAGLWHTVCTSADGDVYAFGG 184
Query: 348 GAYNYGLLGHGNQVSHWVPKRVNGP-LEGIHVSYISCGPWHTAVVTSAGQLFTFGDGTFG 406
+G LG G+ + +P+ ++ P LE ++V ISCG HTA+VT G++F++G +G
Sbjct: 185 N--QFGQLGTGSDQAETLPRLLDCPSLENVNVKTISCGARHTALVTDNGKVFSWGWNKYG 358
Query: 407 ALGHGDRKSVSLPREVESLKGLRTMRASCGVWHTAAVVE 445
LG GD ++P EV +++G +CG WHT + E
Sbjct: 359 QLGLGDVIDRNIPSEV-AIEGCIAKNVACGWWHTLLLAE 472
Score = 87.8 bits (216), Expect = 2e-17
Identities = 45/147 (30%), Positives = 78/147 (52%), Gaps = 2/147 (1%)
Frame = +2
Query: 250 GLGVKIDSLFPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDV 309
G G D L P + S + + ++ +A G H + G+++++G G+LG G D
Sbjct: 47 GQGSTDDELSPTCVSSLLGIQIEGVAAGLWHTVCTSADGDVYAFGGNQFGQLGTGSDQAE 226
Query: 310 LHPKLID--ALSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPK 367
P+L+D +L N N++ ++CG HT VT +G +++WG YG LG G+ + +P
Sbjct: 227 TLPRLLDCPSLENVNVKTISCGARHTALVTDNGKVFSWGWN--KYGQLGLGDVIDRNIPS 400
Query: 368 RVNGPLEGIHVSYISCGPWHTAVVTSA 394
V +EG ++CG WHT ++ +
Sbjct: 401 EV--AIEGCIAKNVACGWWHTLLLAES 475
Score = 83.2 bits (204), Expect = 5e-16
Identities = 55/149 (36%), Positives = 83/149 (54%), Gaps = 3/149 (2%)
Frame = +2
Query: 458 SGKLFTWGDGDKGRLGHGDKEAKLVPTCVA-LVEHNFCQVACGHSLTVALTTSGHVYAMG 516
+G + +G G G+ G G + +L PTCV+ L+ VA G TV + G VYA G
Sbjct: 2 AGAVLAFGWGLYGQCGQGSTDDELSPTCVSSLLGIQIEGVAAGLWHTVCTSADGDVYAFG 181
Query: 517 SPVYGQLGNPQADGK-LPTRVEGKLLKSF-VEEIACGAYHVAVLTLRNEVYTWGKGANGR 574
+GQLG + LP ++ L++ V+ I+CGA H A++T +V++WG G+
Sbjct: 182 GNQFGQLGTGSDQAETLPRLLDCPSLENVNVKTISCGARHTALVTDNGKVFSWGWNKYGQ 361
Query: 575 LGHGDTDDRNNPTLVDALKDKHVKSIACG 603
LG GD DRN P+ V A++ K++ACG
Sbjct: 362 LGLGDVIDRNIPSEV-AIEGCIAKNVACG 445
Score = 80.9 bits (198), Expect = 2e-15
Identities = 53/158 (33%), Positives = 80/158 (50%), Gaps = 4/158 (2%)
Frame = +2
Query: 352 YGLLGHGNQVSHWVPKRVNGPLEGIHVSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHG 411
YG G G+ P V+ L GI + ++ G WHT ++ G ++ FG FG LG G
Sbjct: 35 YGQCGQGSTDDELSPTCVSSLL-GIQIEGVAAGLWHTVCTSADGDVYAFGGNQFGQLGTG 211
Query: 412 DRKSVSLPREVE--SLKGLRTMRASCGVWHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDK 469
++ +LPR ++ SL+ + SCG HTA V + +GK+F+WG
Sbjct: 212 SDQAETLPRLLDCPSLENVNVKTISCGARHTALVTD------------NGKVFSWGWNKY 355
Query: 470 GRLGHGDKEAKLVPTCVALVEHNFCQVACG--HSLTVA 505
G+LG GD + +P+ VA+ VACG H+L +A
Sbjct: 356 GQLGLGDVIDRNIPSEVAIEGCIAKNVACGWWHTLLLA 469
Score = 74.7 bits (182), Expect = 2e-13
Identities = 51/171 (29%), Positives = 79/171 (45%), Gaps = 6/171 (3%)
Frame = +2
Query: 394 AGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGVWHTAAVVEVMVGNSSS 453
AG + FG G +G G G P V SL G++ + G+WHT
Sbjct: 2 AGAVLAFGWGLYGQCGQGSTDDELSPTCVSSLLGIQIEGVAAGLWHTVC----------- 148
Query: 454 SNCSSGKLFTWGDGDKGRLGHGDKEAKLVP---TCVALVEHNFCQVACGHSLTVALTTSG 510
+ G ++ +G G+LG G +A+ +P C +L N ++CG T +T +G
Sbjct: 149 -TSADGDVYAFGGNQFGQLGTGSDQAETLPRLLDCPSLENVNVKTISCGARHTALVTDNG 325
Query: 511 HVYAMGSPVYGQLG-NPQADGKLPTRV--EGKLLKSFVEEIACGAYHVAVL 558
V++ G YGQLG D +P+ V EG + K+ +ACG +H +L
Sbjct: 326 KVFSWGWNKYGQLGLGDVIDRNIPSEVAIEGCIAKN----VACGWWHTLLL 466
Score = 58.2 bits (139), Expect = 2e-08
Identities = 38/100 (38%), Positives = 51/100 (51%), Gaps = 2/100 (2%)
Frame = +2
Query: 236 GDGVVGGGNHRVGSGLGVKIDSLFPKALE--SAVVLDVQNIACGGRHAALVTKQGEIFSW 293
GD GGN G G P+ L+ S ++V+ I+CG RH ALVT G++FSW
Sbjct: 161 GDVYAFGGNQFGQLGTGSDQAETLPRLLDCPSLENVNVKTISCGARHTALVTDNGKVFSW 340
Query: 294 GEESGGRLGHGVDSDVLHPKLIDALSNTNIELVACGEYHT 333
G G+LG G D P + A+ + VACG +HT
Sbjct: 341 GWNKYGQLGLGDVIDRNIPSEV-AIEGCIAKNVACGWWHT 457
>TC87758 similar to PIR|T51387|T51387 UVB-resistance protein-like -
Arabidopsis thaliana, partial (62%)
Length = 894
Score = 98.6 bits (244), Expect = 1e-20
Identities = 83/282 (29%), Positives = 126/282 (44%), Gaps = 14/282 (4%)
Frame = +1
Query: 336 VTLSGDLYTWGNGAYNYGLLGHG-NQVSHWVPKRVNGPLEGIHVSYISCGPWHTAVVTSA 394
+ ++ ++ WG+G G LG G N+ WV P + HV + G ++ + +
Sbjct: 58 ILMASNIIAWGSG--EDGQLGIGTNEDKEWVCSVKALPSQ--HVRSVVAGSRNSLAILND 225
Query: 395 GQLFTFGDGTFGALGH-GDRKSVSLPREVESLKGLRTMRASCGVWHTAAVVEVMVGNSSS 453
G+LFT+G G LGH + K+ ++P +V++L ++A+ G WH AV +
Sbjct: 226 GKLFTWGWNQRGTLGHPAETKTENIPSQVKALSHAHIVQAAIGGWHCLAVDD-------- 381
Query: 454 SNCSSGKLFTWGDGDKGRLGHGD--KEAKLVPTCVALVEHNFC-------QVACGHSLTV 504
GK + WG + G+ G K+ P +V C QVA G + +V
Sbjct: 382 ----HGKAYAWGGNEYGQCGEEPERKDDNGRPLRRDIVIPQPCAPKLSVRQVAAGGTHSV 549
Query: 505 ALTTSGHVYAMGSPVYGQLGNPQADGK---LPTRVEGKLLKSFVEEIACGAYHVAVLTLR 561
LT GHV+ G P P D K +P RV+G ++ IA GA+H L
Sbjct: 550 VLTREGHVWTWGQPW------PPGDIKQISVPVRVQGL---DNIKLIAVGAFHNLALQED 702
Query: 562 NEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACG 603
++ WG G+LG GDT R+ P V L IA G
Sbjct: 703 GTLWAWGNNEYGQLGTGDTQPRSQPVPVQGLSGLTXVXIAXG 828
Score = 87.4 bits (215), Expect = 3e-17
Identities = 71/273 (26%), Positives = 120/273 (43%), Gaps = 13/273 (4%)
Frame = +1
Query: 227 DVFIWGEGTGDGVVGGGNHRVGSGLGVKIDSLFPKALESAVVLDVQNIACGGRHAALVTK 286
++ WG G DG +G +G D + ++++ V+++ G R++ +
Sbjct: 73 NIIAWGSGE-DGQLG---------IGTNEDKEWVCSVKALPSQHVRSVVAGSRNSLAILN 222
Query: 287 QGEIFSWGEESGGRLGHGVDSDVLH-PKLIDALSNTNIELVACGEYHTCAVTLSGDLYTW 345
G++F+WG G LGH ++ + P + ALS+ +I A G +H AV G Y W
Sbjct: 223 DGKLFTWGWNQRGTLGHPAETKTENIPSQVKALSHAHIVQAAIGGWHCLAVDDHGKAYAW 402
Query: 346 GNGAYNYGLLGHGNQVSHWVPKRVNG-PLE-----------GIHVSYISCGPWHTAVVTS 393
G YG G + K NG PL + V ++ G H+ V+T
Sbjct: 403 GGN--EYGQCGEEPE-----RKDDNGRPLRRDIVIPQPCAPKLSVRQVAAGGTHSVVLTR 561
Query: 394 AGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGVWHTAAVVEVMVGNSSS 453
G ++T+G GD K +S+P V+ L ++ + + G +H A+ E
Sbjct: 562 EGHVWTWGQ----PWPPGDIKQISVPVRVQGLDNIKLI--AVGAFHNLALQE-------- 699
Query: 454 SNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCV 486
G L+ WG+ + G+LG GD + + P V
Sbjct: 700 ----DGTLWAWGNNEYGQLGTGDTQPRSQPVPV 786
Score = 42.4 bits (98), Expect = 0.001
Identities = 18/56 (32%), Positives = 33/56 (58%)
Frame = +1
Query: 555 VAVLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAAI 610
++V+ + + + WG G +G+LG G +D+ V AL +HV+S+ G+ + AI
Sbjct: 49 LSVILMASNIIAWGSGEDGQLGIGTNEDKEWVCSVKALPSQHVRSVVAGSRNSLAI 216
>TC88565 similar to GP|20197352|gb|AAC36161.2 expressed protein {Arabidopsis
thaliana}, partial (50%)
Length = 1721
Score = 95.5 bits (236), Expect = 1e-19
Identities = 63/188 (33%), Positives = 95/188 (50%), Gaps = 11/188 (5%)
Frame = +2
Query: 875 EETAKCKAAKEVIKSLTAQLKDMAERLPVGTAKSVKSPSIASFGSNELSFAAIDRLNIQA 934
+E + + K+ IK+LTAQ+KDMA + G K+ K S +S G+ +A D
Sbjct: 350 DEAVQTPSTKQAIKALTAQIKDMAVKAS-GAYKNCKPCSGSSNGNKNKKYADSDM----- 511
Query: 935 TSPEADLTGSNTQLLSNGSSTVSNRSTGQNKQSQSDSTNRNGSRTKDSESRSET------ 988
+D N GS++ + R G+ +++ + R+E+
Sbjct: 512 ---GSDSARFNWAYRRTGSASSTPRMWGKEMEARLKGISSGEGTPTSVSGRTESVVFMEE 682
Query: 989 -----EWVEQDEPGVYITLTSLPGGVIDLKRVRFSRKRFSEKQAENWWAENRVRVYEQYN 1043
EWV Q EPGV IT SLP G DLKR+RFSR+ F++ QA+ WWAEN +V E YN
Sbjct: 683 EDEPKEWVAQVEPGVLITFVSLPEGGNDLKRIRFSREMFNKWQAQRWWAENYDKVMELYN 862
Query: 1044 VRMVDKSS 1051
V+ ++ +
Sbjct: 863 VQRFNQEA 886
Score = 94.4 bits (233), Expect = 2e-19
Identities = 42/78 (53%), Positives = 55/78 (69%)
Frame = +2
Query: 965 KQSQSDSTNRNGSRTKDSESRSETEWVEQDEPGVYITLTSLPGGVIDLKRVRFSRKRFSE 1024
+ S S+ + +G + + S ETEWVEQDEPGVYIT+ +LPGG +L+RVRFSR++F E
Sbjct: 1157 RTSSSEEEDHSGELSISNASDMETEWVEQDEPGVYITIRALPGGTRELRRVRFSREKFGE 1336
Query: 1025 KQAENWWAENRVRVYEQY 1042
A WW ENR R+ EQY
Sbjct: 1337 MHARLWWEENRARIQEQY 1390
>TC87759 similar to PIR|T51387|T51387 UVB-resistance protein-like -
Arabidopsis thaliana, partial (51%)
Length = 976
Score = 93.2 bits (230), Expect = 5e-19
Identities = 56/191 (29%), Positives = 91/191 (47%), Gaps = 15/191 (7%)
Frame = +3
Query: 270 DVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDALSNTNIELVACG 329
+++ IA G H + + G +++WG G+LG G P + LS + +A G
Sbjct: 48 NIKLIAVGAFHNLALQEDGTLWAWGNNEYGQLGTGDTQPRSQPVPVQGLSGLTVVDIAAG 227
Query: 330 EYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSYISCGPWHTA 389
+H+ A+T G++Y WG G + G ++ S +P+RV L G + +SCG H+
Sbjct: 228 GWHSTALTDDGEVYGWGRGEHGRLGFGDSDKSSKMLPQRVQ-LLAGEDIVQVSCGGTHSV 404
Query: 390 VVTSAGQLFTFGDGTFGALGHGDRKSVSLPREV---------------ESLKGLRTMRAS 434
+T G++F+FG G G LG+G + + P EV E+ +
Sbjct: 405 ALTRDGRIFSFGRGDHGRLGYGRKVTTGQPMEVPIDIPPPQNLGDADGEAEGNWIAKLVA 584
Query: 435 CGVWHTAAVVE 445
CG HT A+VE
Sbjct: 585 CGGRHTLAIVE 617
Score = 78.6 bits (192), Expect = 1e-14
Identities = 62/218 (28%), Positives = 97/218 (44%), Gaps = 19/218 (8%)
Frame = +3
Query: 412 DRKSVSLPREVESLKGLRTMRASCGVWHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGR 471
D K +S+P V+ L ++ + + G +H A+ E G L+ WG+ + G+
Sbjct: 3 DIKQISVPVRVQGLDNIKLI--AVGAFHNLALQE------------DGTLWAWGNNEYGQ 140
Query: 472 LGHGDKEAKLVPTCV-ALVEHNFCQVACGHSLTVALTTSGHVYAMGSPVYGQLGNPQADG 530
LG GD + + P V L +A G + ALT G VY G +G+LG +D
Sbjct: 141 LGTGDTQPRSQPVPVQGLSGLTVVDIAAGGWHSTALTDDGEVYGWGRGEHGRLGFGDSDK 320
Query: 531 K---LPTRVEGKLLKSFVEEIACGAYHVAVLTLRNEVYTWGKGANGRLGHGDTDDRNNPT 587
LP RV+ L + +++CG H LT ++++G+G +GRLG+G P
Sbjct: 321 SSKMLPQRVQ-LLAGEDIVQVSCGGTHSVALTRDGRIFSFGRGDHGRLGYGRKVTTGQPM 497
Query: 588 LV---------------DALKDKHVKSIACGTNFTAAI 610
V +A + K +ACG T AI
Sbjct: 498 EVPIDIPPPQNLGDADGEAEGNWIAKLVACGGRHTLAI 611
Score = 73.9 bits (180), Expect = 3e-13
Identities = 52/167 (31%), Positives = 83/167 (49%), Gaps = 18/167 (10%)
Frame = +3
Query: 243 GNHRVGS-GLGVKIDSLFPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRL 301
GN+ G G G P ++ L V +IA GG H+ +T GE++ WG GRL
Sbjct: 120 GNNEYGQLGTGDTQPRSQPVPVQGLSGLTVVDIAAGGWHSTALTDDGEVYGWGRGEHGRL 299
Query: 302 GHGVDSD---VLHPKLIDALSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHG 358
G G DSD + P+ + L+ +I V+CG H+ A+T G ++++G G ++G LG+G
Sbjct: 300 GFG-DSDKSSKMLPQRVQLLAGEDIVQVSCGGTHSVALTRDGRIFSFGRG--DHGRLGYG 470
Query: 359 NQVSHWVPKRV-------------NGPLEGIHVS-YISCGPWHTAVV 391
+V+ P V +G EG ++ ++CG HT +
Sbjct: 471 RKVTTGQPMEVPIDIPPPQNLGDADGEAEGNWIAKLVACGGRHTLAI 611
Score = 61.6 bits (148), Expect = 2e-09
Identities = 39/126 (30%), Positives = 61/126 (47%), Gaps = 15/126 (11%)
Frame = +3
Query: 226 GDVFIWGEGTGDGVVGGGNHRVGSGLGVKIDSLFPKALESAVVLDVQNIACGGRHAALVT 285
G+V+ WG G + R+G G K + P+ ++ D+ ++CGG H+ +T
Sbjct: 258 GEVYGWGRGE--------HGRLGFGDSDKSSKMLPQRVQLLAGEDIVQVSCGGTHSVALT 413
Query: 286 KQGEIFSWGEESGGRLGHG----------VDSDVLHPKLI-----DALSNTNIELVACGE 330
+ G IFS+G GRLG+G V D+ P+ + +A N +LVACG
Sbjct: 414 RDGRIFSFGRGDHGRLGYGRKVTTGQPMEVPIDIPPPQNLGDADGEAEGNWIAKLVACGG 593
Query: 331 YHTCAV 336
HT A+
Sbjct: 594 RHTLAI 611
>TC92196 similar to GP|16604485|gb|AAL24248.1 AT5g11580/F15N18_170
{Arabidopsis thaliana}, partial (34%)
Length = 1213
Score = 85.9 bits (211), Expect = 8e-17
Identities = 86/320 (26%), Positives = 130/320 (39%), Gaps = 49/320 (15%)
Frame = +1
Query: 340 GDLYTWGNGAYNYGLLGHGNQVSHWVPKRVN-----GPLEGIHVSYISCGPWHTAVVTSA 394
G +Y WG G + G LG+G+Q PK++N G + + + I+ G +HT +
Sbjct: 292 GSVYCWGRGMF--GRLGNGSQKDELFPKKLNFGNPNGTQDSVKIVGIAAGSYHTLALAXD 465
Query: 395 GQLFTFGDGTFGALGHGDRK---------------------SVSLPREVESLKGLRTMRA 433
G ++ +G +G LG + + S + P E E L
Sbjct: 466 GSVWCWGYNIYGQLGLCEXEDSHDSLVPCLLSRFLELQPHNSSTDPSEAEDKASLEMCTV 645
Query: 434 SCGVWHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHG-DKEAKLVPTCV-ALVEH 491
G G S + + G L+ WG+ + G + PT V
Sbjct: 646 KAG------------GMMSLAIDNRGTLWMWGNIPQESKESGISLVSSFTPTPVWDFHGR 789
Query: 492 NFCQVACGHSLTVALTTS--GH------VYAMGSPVYGQLGNPQADGKLPTRVEGKLLKS 543
+VACG+ VAL + H Y+ G +GQLG +L V K+L
Sbjct: 790 TVVKVACGNEHVVALVNAIDSHEDEDLLCYSWGYNSHGQLGLGDRQSRLHPEVV-KILDE 966
Query: 544 ----FVEEIACGAYHVAVL--------TLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDA 591
+ E+ACGA+H A+L TL + +T+G G NG+LGHG T PT V
Sbjct: 967 ESPWTIYEVACGAFHTALLAHKKKPDATLESMCWTFGLGENGQLGHGTTQSTLFPTPVKE 1146
Query: 592 LKDK-HVKSIACGTNFTAAI 610
L H+ S+ CG T+ +
Sbjct: 1147LPQSIHLISVDCGLFHTSVV 1206
Score = 80.5 bits (197), Expect = 3e-15
Identities = 73/312 (23%), Positives = 123/312 (39%), Gaps = 38/312 (12%)
Frame = +1
Query: 286 KQGEIFSWGEESGGRLGHGVDSDVLHPKLID------ALSNTNIELVACGEYHTCAVTLS 339
+ G ++ WG GRLG+G D L PK ++ + I +A G YHT A+
Sbjct: 286 EDGSVYCWGRGMFGRLGNGSQKDELFPKKLNFGNPNGTQDSVKIVGIAAGSYHTLALAXD 465
Query: 340 GDLYTWGNGAYNY-GLLGHGNQVSHWVPKRVNGPLE-----------------GIHVSYI 381
G ++ WG Y GL + VP ++ LE + + +
Sbjct: 466 GSVWCWGYNIYGQLGLCEXEDSHDSLVPCLLSRFLELQPHNSSTDPSEAEDKASLEMCTV 645
Query: 382 SCGPWHTAVVTSAGQLFTFGDGTFGALGHG-DRKSVSLPREVESLKGLRTMRASCGVWHT 440
G + + + G L+ +G+ + G S P V G ++ +CG H
Sbjct: 646 KAGGMMSLAIDNRGTLWMWGNIPQESKESGISLVSSFTPTPVWDFHGRTVVKVACGNEHV 825
Query: 441 AAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCVALVEH----NFCQV 496
A+V + + ++WG G+LG GD++++L P V +++ +V
Sbjct: 826 VALVNAIDSHEDEDLLC----YSWGYNSHGQLGLGDRQSRLHPEVVKILDEESPWTIYEV 993
Query: 497 ACGHSLTVAL--------TTSGHVYAMGSPVYGQLGNPQADGKL-PTRVEGKLLKSFVEE 547
ACG T L T + G GQLG+ L PT V+ +
Sbjct: 994 ACGAFHTALLAHKKKPDATLESMCWTFGLGENGQLGHGTTQSTLFPTPVKELPQSIHLIS 1173
Query: 548 IACGAYHVAVLT 559
+ CG +H +V++
Sbjct: 1174VDCGLFHTSVVS 1209
Score = 68.2 bits (165), Expect = 2e-11
Identities = 50/142 (35%), Positives = 68/142 (47%), Gaps = 19/142 (13%)
Frame = +1
Query: 271 VQNIACGGRHA-----ALVTKQGE---IFSWGEESGGRLGHGVDSDVLHP---KLIDALS 319
V +ACG H A+ + + E +SWG S G+LG G LHP K++D S
Sbjct: 793 VVKVACGNEHVVALVNAIDSHEDEDLLCYSWGYNSHGQLGLGDRQSRLHPEVVKILDEES 972
Query: 320 NTNIELVACGEYHTCAV--------TLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNG 371
I VACG +HT + TL +T+G G G LGHG S P V
Sbjct: 973 PWTIYEVACGAFHTALLAHKKKPDATLESMCWTFGLG--ENGQLGHGTTQSTLFPTPVKE 1146
Query: 372 PLEGIHVSYISCGPWHTAVVTS 393
+ IH+ + CG +HT+VV+S
Sbjct: 1147LPQSIHLISVDCGLFHTSVVSS 1212
>TC81917 similar to GP|10176914|dbj|BAB10107. contains similarity to
regulator of chromosome condensation~gene_id:MAE1.11
{Arabidopsis thaliana}, partial (29%)
Length = 394
Score = 74.3 bits (181), Expect = 2e-13
Identities = 44/135 (32%), Positives = 68/135 (49%), Gaps = 1/135 (0%)
Frame = +2
Query: 408 LGHGDRKSVSLPREVESLKGLRTMRASCGVWHTAAVVEVMVGNSSSSNCSSGKLFTWGDG 467
LG GDR PR V SL+ +R ++ + G +H+ A+ + GK+ +WG G
Sbjct: 11 LGRGDRIGGWRPRPVPSLEKVRVIQIASGGYHSLALTD------------DGKVLSWGHG 154
Query: 468 DKGRLGHGDKEAKLVPTCVALVEH-NFCQVACGHSLTVALTTSGHVYAMGSPVYGQLGNP 526
+G+LGHG E + +PT V + H + ++CG + + A+T G +Y G+ QLG P
Sbjct: 155 GQGQLGHGSVENQKIPTLVEAIAHEHIIYISCGGASSAAVTDEGKLYMWGNASDSQLGVP 334
Query: 527 QADGKLPTRVEGKLL 541
P VE L
Sbjct: 335 GLPAIQPYPVEVNFL 379
Score = 72.4 bits (176), Expect = 9e-13
Identities = 36/100 (36%), Positives = 56/100 (56%)
Frame = +2
Query: 250 GLGVKIDSLFPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDV 309
G G +I P+ + S + V IA GG H+ +T G++ SWG G+LGHG +
Sbjct: 14 GRGDRIGGWRPRPVPSLEKVRVIQIASGGYHSLALTDDGKVLSWGHGGQGQLGHGSVENQ 193
Query: 310 LHPKLIDALSNTNIELVACGEYHTCAVTLSGDLYTWGNGA 349
P L++A+++ +I ++CG + AVT G LY WGN +
Sbjct: 194 KIPTLVEAIAHEHIIYISCGGASSAAVTDEGKLYMWGNAS 313
Score = 65.5 bits (158), Expect = 1e-10
Identities = 37/106 (34%), Positives = 55/106 (50%), Gaps = 4/106 (3%)
Frame = +2
Query: 522 QLGNPQADGKLPTRVEGKLLKSFVEEIACGAYHVAVLTLRNEVYTWGKGANGRLGHGDTD 581
+LG G R L K V +IA G YH LT +V +WG G G+LGHG +
Sbjct: 8 ELGRGDRIGGWRPRPVPSLEKVRVIQIASGGYHSLALTDDGKVLSWGHGGQGQLGHGSVE 187
Query: 582 DRNNPTLVDALKDKHVKSIACGTNFTAAIC----LHKWVSGVDQSM 623
++ PTLV+A+ +H+ I+CG +AA+ L+ W + D +
Sbjct: 188 NQKIPTLVEAIAHEHIIYISCGGASSAAVTDEGKLYMWGNASDSQL 325
Score = 65.5 bits (158), Expect = 1e-10
Identities = 37/119 (31%), Positives = 61/119 (51%)
Frame = +2
Query: 355 LGHGNQVSHWVPKRVNGPLEGIHVSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRK 414
LG G+++ W P+ V LE + V I+ G +H+ +T G++ ++G G G LGHG +
Sbjct: 11 LGRGDRIGGWRPRPVPS-LEKVRVIQIASGGYHSLALTDDGKVLSWGHGGQGQLGHGSVE 187
Query: 415 SVSLPREVESLKGLRTMRASCGVWHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLG 473
+ +P VE++ + SCG G SS++ GKL+ WG+ +LG
Sbjct: 188 NQKIPTLVEAIAHEHIIYISCG------------GASSAAVTDEGKLYMWGNASDSQLG 328
Score = 63.5 bits (153), Expect = 4e-10
Identities = 36/109 (33%), Positives = 57/109 (52%)
Frame = +2
Query: 301 LGHGVDSDVLHPKLIDALSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQ 360
LG G P+ + +L + +A G YH+ A+T G + +WG+G G LGHG+
Sbjct: 11 LGRGDRIGGWRPRPVPSLEKVRVIQIASGGYHSLALTDDGKVLSWGHG--GQGQLGHGSV 184
Query: 361 VSHWVPKRVNGPLEGIHVSYISCGPWHTAVVTSAGQLFTFGDGTFGALG 409
+ +P V + H+ YISCG +A VT G+L+ +G+ + LG
Sbjct: 185 ENQKIPTLVEA-IAHEHIIYISCGGASSAAVTDEGKLYMWGNASDSQLG 328
Score = 50.8 bits (120), Expect = 3e-06
Identities = 35/107 (32%), Positives = 53/107 (48%), Gaps = 2/107 (1%)
Frame = +2
Query: 472 LGHGDKEAKLVPTCVALVEH-NFCQVACGHSLTVALTTSGHVYAMGSPVYGQLGNPQADG 530
LG GD+ P V +E Q+A G ++ALT G V + G GQLG+ +
Sbjct: 11 LGRGDRIGGWRPRPVPSLEKVRVIQIASGGYHSLALTDDGKVLSWGHGGQGQLGHGSVEN 190
Query: 531 -KLPTRVEGKLLKSFVEEIACGAYHVAVLTLRNEVYTWGKGANGRLG 576
K+PT VE + + I+CG A +T ++Y WG ++ +LG
Sbjct: 191 QKIPTLVEA-IAHEHIIYISCGGASSAAVTDEGKLYMWGNASDSQLG 328
Score = 42.0 bits (97), Expect = 0.001
Identities = 24/77 (31%), Positives = 37/77 (47%)
Frame = +2
Query: 226 GDVFIWGEGTGDGVVGGGNHRVGSGLGVKIDSLFPKALESAVVLDVQNIACGGRHAALVT 285
G V WG G G G +G G+ + P +E+ + I+CGG +A VT
Sbjct: 128 GKVLSWGHG-GQGQLGHGSVE---------NQKIPTLVEAIAHEHIIYISCGGASSAAVT 277
Query: 286 KQGEIFSWGEESGGRLG 302
+G+++ WG S +LG
Sbjct: 278 DEGKLYMWGNASDSQLG 328
>AL386293 weakly similar to GP|4079809|gb| HERC2 {Homo sapiens}, partial (1%)
Length = 528
Score = 65.5 bits (158), Expect = 1e-10
Identities = 44/107 (41%), Positives = 58/107 (54%), Gaps = 8/107 (7%)
Frame = +3
Query: 260 PKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLG--HGVDS--DVLHPKLI 315
P A+E L V+ IACGG H+A++T G+++++G GRLG G DS + P LI
Sbjct: 84 PTAIEFFQGLRVKEIACGGFHSAVITDSGDLYTFGWNHFGRLGISSGKDSMMNCAEPSLI 263
Query: 316 DALSNTNIEL----VACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHG 358
+ +IEL VACG HT A+T D W G YG LG G
Sbjct: 264 EFGEENDIELNVLKVACGSAHTVAIT--DDYRLWSCGWGKYGQLGLG 398
Score = 53.5 bits (127), Expect = 4e-07
Identities = 37/127 (29%), Positives = 60/127 (47%), Gaps = 9/127 (7%)
Frame = +3
Query: 407 ALGHGDRKSVSLPREVESLKGLRTMRASCGVWHTAAVVEVMVGNSSSSNCSSGKLFTWGD 466
+L G + P +E +GLR +CG +H+A + + SG L+T+G
Sbjct: 48 SLDMGIYNLIRKPTAIEFFQGLRVKEIACGGFHSAVITD------------SGDLYTFGW 191
Query: 467 GDKGRLGHGDKEAKLV----PTCVAL-----VEHNFCQVACGHSLTVALTTSGHVYAMGS 517
GRLG + ++ P+ + +E N +VACG + TVA+T +++ G
Sbjct: 192 NHFGRLGISSGKDSMMNCAEPSLIEFGEENDIELNVLKVACGSAHTVAITDDYRLWSCGW 371
Query: 518 PVYGQLG 524
YGQLG
Sbjct: 372 GKYGQLG 392
Score = 52.8 bits (125), Expect(2) = 1e-10
Identities = 34/116 (29%), Positives = 59/116 (50%), Gaps = 5/116 (4%)
Frame = +3
Query: 301 LGHGVDSDVLHPKLIDALSNTNIELVACGEYHTCAVTLSGDLYTWG-NGAYNYGL-LGHG 358
L G+ + + P I+ ++ +ACG +H+ +T SGDLYT+G N G+ G
Sbjct: 51 LDMGIYNLIRKPTAIEFFQGLRVKEIACGGFHSAVITDSGDLYTFGWNHFGRLGISSGKD 230
Query: 359 NQVSHWVPKRVNGPLEG---IHVSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHG 411
+ ++ P + E ++V ++CG HT +T +L++ G G +G LG G
Sbjct: 231 SMMNCAEPSLIEFGEENDIELNVLKVACGSAHTVAITDDYRLWSCGWGKYGQLGLG 398
Score = 51.6 bits (122), Expect(2) = 4e-09
Identities = 34/110 (30%), Positives = 52/110 (46%), Gaps = 8/110 (7%)
Frame = +3
Query: 374 EGIHVSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESL-------- 425
+G+ V I+CG +H+AV+T +G L+TFG FG LG K + SL
Sbjct: 105 QGLRVKEIACGGFHSAVITDSGDLYTFGWNHFGRLGISSGKDSMMNCAEPSLIEFGEEND 284
Query: 426 KGLRTMRASCGVWHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHG 475
L ++ +CG HT A+ + +L++ G G G+LG G
Sbjct: 285 IELNVLKVACGSAHTVAITD------------DYRLWSCGWGKYGQLGLG 398
Score = 44.7 bits (104), Expect = 2e-04
Identities = 27/74 (36%), Positives = 41/74 (54%), Gaps = 8/74 (10%)
Frame = +3
Query: 545 VEEIACGAYHVAVLTLRNEVYTWGKGANGRLGHGDTDDR----NNPTLVDALKDK----H 596
V+EIACG +H AV+T ++YT+G GRLG D P+L++ ++ +
Sbjct: 117 VKEIACGGFHSAVITDSGDLYTFGWNHFGRLGISSGKDSMMNCAEPSLIEFGEENDIELN 296
Query: 597 VKSIACGTNFTAAI 610
V +ACG+ T AI
Sbjct: 297 VLKVACGSAHTVAI 338
Score = 32.7 bits (73), Expect(2) = 1e-10
Identities = 12/20 (60%), Positives = 15/20 (75%)
Frame = +2
Query: 285 TKQGEIFSWGEESGGRLGHG 304
T+ GE+FSWG G+LGHG
Sbjct: 2 TRDGEVFSWGSGRFGQLGHG 61
Score = 31.6 bits (70), Expect = 1.7
Identities = 11/18 (61%), Positives = 15/18 (83%)
Frame = +2
Query: 459 GKLFTWGDGDKGRLGHGD 476
G++F+WG G G+LGHGD
Sbjct: 11 GEVFSWGSGRFGQLGHGD 64
Score = 31.6 bits (70), Expect = 1.7
Identities = 12/21 (57%), Positives = 15/21 (71%)
Frame = +2
Query: 559 TLRNEVYTWGKGANGRLGHGD 579
T EV++WG G G+LGHGD
Sbjct: 2 TRDGEVFSWGSGRFGQLGHGD 64
Score = 28.5 bits (62), Expect(2) = 4e-09
Identities = 11/27 (40%), Positives = 18/27 (65%)
Frame = +2
Query: 337 TLSGDLYTWGNGAYNYGLLGHGNQVSH 363
T G++++WG+G +G LGHG+ H
Sbjct: 2 TRDGEVFSWGSG--RFGQLGHGDLQPH 76
>TC91034 similar to GP|20161135|dbj|BAB90063. hypothetical protein {Oryza
sativa (japonica cultivar-group)}, partial (16%)
Length = 639
Score = 61.6 bits (148), Expect = 2e-09
Identities = 38/104 (36%), Positives = 53/104 (50%), Gaps = 3/104 (2%)
Frame = +3
Query: 271 VQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDALSNTNIELV--AC 328
V +A G H L+ G ++SWG + GRLG G + D L P + S +I +V A
Sbjct: 108 VTAVAAGEAHTLLLAGDGRVYSWGRGTLGRLGLGSEQDQLFPAQVKFGSQESIRIVGIAA 287
Query: 329 GEYHTCAVTLSGDLYTWGNGAYN-YGLLGHGNQVSHWVPKRVNG 371
G YH+ A+ G ++ WG YN YG LG N+ PK +G
Sbjct: 288 GAYHSLALADDGSVWCWG---YNLYGQLGIINEEDAHNPKYTDG 410
Score = 51.2 bits (121), Expect = 2e-06
Identities = 33/97 (34%), Positives = 47/97 (48%), Gaps = 2/97 (2%)
Frame = +3
Query: 496 VACGHSLTVALTTSGHVYAMGSPVYGQLG-NPQADGKLPTRVEGKLLKSF-VEEIACGAY 553
VA G + T+ L G VY+ G G+LG + D P +V+ +S + IA GAY
Sbjct: 117 VAAGEAHTLLLAGDGRVYSWGRGTLGRLGLGSEQDQLFPAQVKFGSQESIRIVGIAAGAY 296
Query: 554 HVAVLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVD 590
H L V+ WG G+LG + +D +NP D
Sbjct: 297 HSLALADDGSVWCWGYNLYGQLGIINEEDAHNPKYTD 407
Score = 44.7 bits (104), Expect = 2e-04
Identities = 24/69 (34%), Positives = 37/69 (52%), Gaps = 3/69 (4%)
Frame = +3
Query: 459 GKLFTWGDGDKGRLGHGDKEAKLVPTCVALVEHNFCQ---VACGHSLTVALTTSGHVYAM 515
G++++WG G GRLG G ++ +L P V + +A G ++AL G V+
Sbjct: 159 GRVYSWGRGTLGRLGLGSEQDQLFPAQVKFGSQESIRIVGIAAGAYHSLALADDGSVWCW 338
Query: 516 GSPVYGQLG 524
G +YGQLG
Sbjct: 339 GYNLYGQLG 365
Score = 38.1 bits (87), Expect = 0.018
Identities = 22/68 (32%), Positives = 32/68 (46%), Gaps = 2/68 (2%)
Frame = +3
Query: 545 VEEIACGAYHVAVLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVK--SIAC 602
V +A G H +L VY+WG+G GRLG G D+ P V + ++ IA
Sbjct: 108 VTAVAAGEAHTLLLAGDGRVYSWGRGTLGRLGLGSEQDQLFPAQVKFGSQESIRIVGIAA 287
Query: 603 GTNFTAAI 610
G + A+
Sbjct: 288 GAYHSLAL 311
>BF644238 similar to GP|10176914|dbj contains similarity to regulator of
chromosome condensation~gene_id:MAE1.11 {Arabidopsis
thaliana}, partial (16%)
Length = 321
Score = 39.3 bits (90), Expect = 0.008
Identities = 20/51 (39%), Positives = 27/51 (52%)
Frame = +1
Query: 434 SCGVWHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPT 484
SCG++H+ VV+ G L+ WG GD GRLG G + + VPT
Sbjct: 76 SCGLFHSCLVVD-------------GGLWVWGKGDGGRLGLGHESSMFVPT 189
Score = 38.9 bits (89), Expect = 0.011
Identities = 25/65 (38%), Positives = 34/65 (51%)
Frame = +1
Query: 537 EGKLLKSFVEEIACGAYHVAVLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKH 596
EG K I+CG +H + L + ++ WGKG GRLG G PTL L ++
Sbjct: 40 EGSNNKGVEVGISCGLFH-SCLVVDGGLWVWGKGDGGRLGLGHESSMFVPTLNPHL--EN 210
Query: 597 VKSIA 601
VKS+A
Sbjct: 211 VKSVA 225
Score = 38.9 bits (89), Expect(2) = 5e-08
Identities = 17/32 (53%), Positives = 23/32 (71%)
Frame = +2
Query: 379 SYISCGPWHTAVVTSAGQLFTFGDGTFGALGH 410
S + G H+ +TSAG++FT+G G FGALGH
Sbjct: 215 SLLLLGGLHSVALTSAGEVFTWGYGGFGALGH 310
Score = 37.4 bits (85), Expect(2) = 5e-08
Identities = 22/59 (37%), Positives = 37/59 (62%), Gaps = 1/59 (1%)
Frame = +1
Query: 319 SNTNIEL-VACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGI 376
+N +E+ ++CG +H+C V + G L+ WG G + G LG G++ S +VP +N LE +
Sbjct: 49 NNKGVEVGISCGLFHSCLV-VDGGLWVWGKG--DGGRLGLGHESSMFVP-TLNPHLENV 213
Score = 37.0 bits (84), Expect = 0.040
Identities = 20/47 (42%), Positives = 27/47 (56%)
Frame = +1
Query: 274 IACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDALSN 320
I+CG H+ LV G ++ WG+ GGRLG G +S + P L L N
Sbjct: 73 ISCGLFHSCLVV-DGGLWVWGKGDGGRLGLGHESSMFVPTLNPHLEN 210
Score = 35.8 bits (81), Expect = 0.090
Identities = 14/29 (48%), Positives = 18/29 (61%)
Frame = +2
Query: 277 GGRHAALVTKQGEIFSWGEESGGRLGHGV 305
GG H+ +T GE+F+WG G LGH V
Sbjct: 230 GGLHSVALTSAGEVFTWGYGGFGALGHSV 316
Score = 33.5 bits (75), Expect = 0.45
Identities = 14/27 (51%), Positives = 15/27 (54%)
Frame = +2
Query: 551 GAYHVAVLTLRNEVYTWGKGANGRLGH 577
G H LT EV+TWG G G LGH
Sbjct: 230 GGLHSVALTSAGEVFTWGYGGFGALGH 310
Score = 29.3 bits (64), Expect = 8.4
Identities = 13/26 (50%), Positives = 17/26 (65%)
Frame = +2
Query: 449 GNSSSSNCSSGKLFTWGDGDKGRLGH 474
G S + S+G++FTWG G G LGH
Sbjct: 233 GLHSVALTSAGEVFTWGYGGFGALGH 310
>BG454336 similar to GP|13548329|em putative protein {Arabidopsis thaliana},
partial (25%)
Length = 663
Score = 43.9 bits (102), Expect = 3e-04
Identities = 22/68 (32%), Positives = 37/68 (54%), Gaps = 2/68 (2%)
Frame = +3
Query: 271 VQNIACGGRHAALVTKQGEIFSWGEESGGRLGHG--VDSDVLHPKLIDALSNTNIELVAC 328
V I+ G H++ +T GE++ WG+ + G+LG G + V P ++ L IE+ A
Sbjct: 456 VVRISAGYNHSSAITVDGELYMWGKNTTGQLGLGKKAPNIVPLPTKVEYLDGITIEMAAL 635
Query: 329 GEYHTCAV 336
G H+ A+
Sbjct: 636 GSEHSLAI 659
Score = 37.7 bits (86), Expect(2) = 1e-07
Identities = 22/58 (37%), Positives = 31/58 (52%), Gaps = 3/58 (5%)
Frame = +2
Query: 453 SSNCSSGKLFT--WGDGDKGRLGHGDKEAKLVPT-CVALVEHNFCQVACGHSLTVALT 507
S + GK F WG+GD GRLG G+ ++ P C + N +ACG + T+ LT
Sbjct: 248 SFSSEPGKRFAALWGNGDYGRLGLGNLNSQWKPAICTSFHNQNVQAIACGGAHTLFLT 421
Score = 37.4 bits (85), Expect(2) = 1e-07
Identities = 26/82 (31%), Positives = 40/82 (48%), Gaps = 2/82 (2%)
Frame = +3
Query: 531 KLPTRVEGKLLKSFVEEIACGAYHVAVLTLRNEVYTWGKGANGRLGHGDTDDR--NNPTL 588
++P +V G K V I+ G H + +T+ E+Y WGK G+LG G PT
Sbjct: 420 QVPLQVFGHEKK--VVRISAGYNHSSAITVDGELYMWGKNTTGQLGLGKKAPNIVPLPTK 593
Query: 589 VDALKDKHVKSIACGTNFTAAI 610
V+ L ++ A G+ + AI
Sbjct: 594 VEYLDGITIEMAALGSEHSLAI 659
Score = 37.0 bits (84), Expect(2) = 8e-07
Identities = 22/54 (40%), Positives = 27/54 (49%)
Frame = +2
Query: 345 WGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSYISCGPWHTAVVTSAGQLF 398
WGNG +YG LG GN S W P + +V I+CG HT +T A F
Sbjct: 287 WGNG--DYGRLGLGNLNSQWKP-AICTSFHNQNVQAIACGGAHTLFLTGATSSF 439
Score = 35.8 bits (81), Expect(2) = 0.001
Identities = 20/67 (29%), Positives = 33/67 (48%), Gaps = 1/67 (1%)
Frame = +3
Query: 326 VACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGP-LEGIHVSYISCG 384
++ G H+ A+T+ G+LY WG G LG G + + VP L+GI + + G
Sbjct: 465 ISAGYNHSSAITVDGELYMWGKN--TTGQLGLGKKAPNIVPLPTKVEYLDGITIEMAALG 638
Query: 385 PWHTAVV 391
H+ +
Sbjct: 639 SEHSLAI 659
Score = 35.4 bits (80), Expect = 0.12
Identities = 14/37 (37%), Positives = 24/37 (64%)
Frame = +2
Query: 567 WGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACG 603
WG G GRLG G+ + + P + + +++V++IACG
Sbjct: 287 WGNGDYGRLGLGNLNSQWKPAICTSFHNQNVQAIACG 397
Score = 35.4 bits (80), Expect = 0.12
Identities = 16/45 (35%), Positives = 23/45 (50%)
Frame = +2
Query: 293 WGEESGGRLGHGVDSDVLHPKLIDALSNTNIELVACGEYHTCAVT 337
WG GRLG G + P + + N N++ +ACG HT +T
Sbjct: 287 WGNGDYGRLGLGNLNSQWKPAICTSFHNQNVQAIACGGAHTLFLT 421
Score = 35.0 bits (79), Expect(2) = 8e-07
Identities = 18/46 (39%), Positives = 24/46 (52%), Gaps = 1/46 (2%)
Frame = +3
Query: 439 HTAAVVEVMVG-NSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVP 483
H VV + G N SS+ G+L+ WG G+LG G K +VP
Sbjct: 444 HEKKVVRISAGYNHSSAITVDGELYMWGKNTTGQLGLGKKAPNIVP 581
Score = 25.4 bits (54), Expect(2) = 0.001
Identities = 13/28 (46%), Positives = 15/28 (53%)
Frame = +2
Query: 270 DVQNIACGGRHAALVTKQGEIFSWGEES 297
+VQ IACGG H +T F EES
Sbjct: 374 NVQAIACGGAHTLFLTGATSSFWAREES 457
>BG644504 similar to PIR|A86443|A864 probable major intrinsic protein
[imported] - Arabidopsis thaliana, partial (26%)
Length = 728
Score = 52.8 bits (125), Expect = 7e-07
Identities = 31/74 (41%), Positives = 40/74 (53%)
Frame = +2
Query: 952 GSSTVSNRSTGQNKQSQSDSTNRNGSRTKDSESRSETEWVEQDEPGVYITLTSLPGGVID 1011
GSS +R+T ++ S S + S E+EWVEQDEPGVYIT+ L G +
Sbjct: 104 GSSMEPSRATTSSRDEPSIS----------NASEMESEWVEQDEPGVYITIRQLADGTRE 253
Query: 1012 LKRVRFSRKRFSEK 1025
L+RVRF S K
Sbjct: 254 LRRVRFRYYNASSK 295
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.313 0.130 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 32,634,440
Number of Sequences: 36976
Number of extensions: 514517
Number of successful extensions: 2932
Number of sequences better than 10.0: 88
Number of HSP's better than 10.0 without gapping: 2593
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2794
length of query: 1061
length of database: 9,014,727
effective HSP length: 106
effective length of query: 955
effective length of database: 5,095,271
effective search space: 4865983805
effective search space used: 4865983805
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146692.2


