

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146683.17 - phase: 0
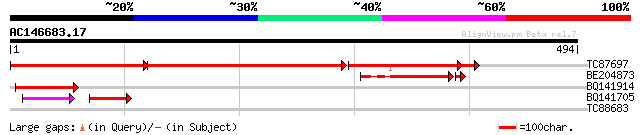
(494 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC87697 similar to PIR|T47802|T47802 hypothetical protein F24G16... 328 0.0
BE204873 similar to GP|18767124|gb| unknown {Saccharomyces cerev... 105 2e-24
BQ141914 64 1e-10
BQ141705 44 1e-06
TC88683 similar to GP|21593856|gb|AAM65823.1 unknown {Arabidopsi... 28 6.2
>TC87697 similar to PIR|T47802|T47802 hypothetical protein F24G16.50 -
Arabidopsis thaliana, partial (7%)
Length = 1514
Score = 328 bits (841), Expect(4) = 0.0
Identities = 172/173 (99%), Positives = 172/173 (99%)
Frame = +1
Query: 121 GINETFSDSINKGENALRSSVDTATSFIDSVVKNATTSADNAFSKVFSAADQTGDLANKK 180
GINETFSDSINKGENALRSSVDTATSFIDSV KNATTSADNAFSKVFSAADQTGDLANKK
Sbjct: 634 GINETFSDSINKGENALRSSVDTATSFIDSVAKNATTSADNAFSKVFSAADQTGDLANKK 813
Query: 181 ITSFSSEIDGVTSKAPGLVIDVLRRTVVAVESSLSSGASYVVYLYGSAKEFIPAEIRDTV 240
ITSFSSEIDGVTSKAPGLVIDVLRRTVVAVESSLSSGASYVVYLYGSAKEFIPAEIRDTV
Sbjct: 814 ITSFSSEIDGVTSKAPGLVIDVLRRTVVAVESSLSSGASYVVYLYGSAKEFIPAEIRDTV 993
Query: 241 NIYEDKAAQVLRPVGSATQQIYMAFYSLEKSLGLDPNDPIIPFVVFVGSSATL 293
NIYEDKAAQVLRPVGSATQQIYMAFYSLEKSLGLDPNDPIIPFVVFVGSSATL
Sbjct: 994 NIYEDKAAQVLRPVGSATQQIYMAFYSLEKSLGLDPNDPIIPFVVFVGSSATL 1152
Score = 235 bits (600), Expect(4) = 0.0
Identities = 121/121 (100%), Positives = 121/121 (100%)
Frame = +3
Query: 1 DFESRGVVEENVHLGLYHGTPSLRSSFTAQAIKTVSAWDNSSLSTGEFRYSLSTVPEELV 60
DFESRGVVEENVHLGLYHGTPSLRSSFTAQAIKTVSAWDNSSLSTGEFRYSLSTVPEELV
Sbjct: 273 DFESRGVVEENVHLGLYHGTPSLRSSFTAQAIKTVSAWDNSSLSTGEFRYSLSTVPEELV 452
Query: 61 DFAKQSTEVSEPLVAPTVHPEAILSSTDITSGKIESVPSLIKGGNESLAATKASATEVFA 120
DFAKQSTEVSEPLVAPTVHPEAILSSTDITSGKIESVPSLIKGGNESLAATKASATEVFA
Sbjct: 453 DFAKQSTEVSEPLVAPTVHPEAILSSTDITSGKIESVPSLIKGGNESLAATKASATEVFA 632
Query: 121 G 121
G
Sbjct: 633 G 635
Score = 174 bits (442), Expect(4) = 0.0
Identities = 94/107 (87%), Positives = 95/107 (87%), Gaps = 8/107 (7%)
Frame = +2
Query: 296 QIYAGLFIGCGNMEVTQEIYPLNQLSSSWPGTQML--------TCEKKMVFLIFDGLLGF 347
Q + GLFIGCGNMEVTQEIYPLNQLSSSWPGTQML TCEKKMVFLIFDGLLGF
Sbjct: 1142 QQHYGLFIGCGNMEVTQEIYPLNQLSSSWPGTQMLYL*MSAPRTCEKKMVFLIFDGLLGF 1321
Query: 348 AMQV*LQLRLMAPYASC*RGVET*MTP*LLLLFKI*KLLRTVPRLLC 394
AMQV*LQLRLMAPYASC*RGVET*MTP*LLLLFKI*KLLRTVPR C
Sbjct: 1322 AMQV*LQLRLMAPYASC*RGVET*MTP*LLLLFKI*KLLRTVPRGYC 1462
Score = 29.6 bits (65), Expect(4) = 0.0
Identities = 16/19 (84%), Positives = 16/19 (84%), Gaps = 1/19 (5%)
Frame = +1
Query: 392 LLCWML-MVLVLKALQDP* 409
LLCW MVLVLKALQDP*
Sbjct: 1456 LLCWDAEMVLVLKALQDP* 1512
>BE204873 similar to GP|18767124|gb| unknown {Saccharomyces cerevisiae},
partial (30%)
Length = 574
Score = 105 bits (262), Expect(2) = 2e-24
Identities = 64/81 (79%), Positives = 66/81 (81%)
Frame = +2
Query: 306 GNMEVTQEIYPLNQLSSSWPGTQMLTCEKKMVFLIFDGLLGFAMQV*LQLRLMAPYASC* 365
G M+ TQ +Y L S P T CEKKMVFLIFDGLLGFAMQV*LQLRLMAPYASC*
Sbjct: 326 GKMKRTQMLY----L*MSAPRT----CEKKMVFLIFDGLLGFAMQV*LQLRLMAPYASC* 481
Query: 366 RGVET*MTP*LLLLFKI*KLL 386
RGVET*MTP*LLLLFKI*KLL
Sbjct: 482 RGVET*MTP*LLLLFKI*KLL 544
Score = 25.0 bits (53), Expect(2) = 2e-24
Identities = 9/9 (100%), Positives = 9/9 (100%)
Frame = +3
Query: 389 VPRLLCWML 397
VPRLLCWML
Sbjct: 546 VPRLLCWML 572
>BQ141914
Length = 810
Score = 63.9 bits (154), Expect = 1e-10
Identities = 33/55 (60%), Positives = 40/55 (72%)
Frame = +1
Query: 6 GVVEENVHLGLYHGTPSLRSSFTAQAIKTVSAWDNSSLSTGEFRYSLSTVPEELV 60
GV ++N H G +HGTPSL SSFTAQAIKTV DN+SL TG+ SL TVP ++
Sbjct: 154 GVAQQNAH*G*HHGTPSLISSFTAQAIKTVLYRDNTSLHTGQSTLSLFTVPR*II 318
>BQ141705
Length = 740
Score = 43.9 bits (102), Expect(2) = 1e-06
Identities = 19/37 (51%), Positives = 28/37 (75%)
Frame = +1
Query: 70 SEPLVAPTVHPEAILSSTDITSGKIESVPSLIKGGNE 106
++P++ PT HPEA+L ++DI +GKI P+L GGNE
Sbjct: 415 NDPIIPPTEHPEALLFTSDINTGKIAYHPTLN*GGNE 525
Score = 26.6 bits (57), Expect(2) = 1e-06
Identities = 15/45 (33%), Positives = 22/45 (48%)
Frame = +3
Query: 12 VHLGLYHGTPSLRSSFTAQAIKTVSAWDNSSLSTGEFRYSLSTVP 56
VH+ GTP LR + + +T WD LST + Y ++ P
Sbjct: 237 VHMR*CGGTP*LRYKCSGKL*RTGGNWDIP*LSTEQLTYIITPAP 371
>TC88683 similar to GP|21593856|gb|AAM65823.1 unknown {Arabidopsis
thaliana}, partial (77%)
Length = 673
Score = 28.5 bits (62), Expect = 6.2
Identities = 17/78 (21%), Positives = 36/78 (45%), Gaps = 2/78 (2%)
Frame = -2
Query: 210 VESSLSSGASYVVYLYGSAKEFIPAEIRDTVNIYEDKAAQVLRPVGSATQQIYMAFYS-- 267
VE L+ S + + + F+P + +NI + ++P+GS I M ++
Sbjct: 453 VEDPLNRSFSNNILIIPNKINFLPNITKKIINILLTQIVSTIKPIGSKNTHIKMKPFASV 274
Query: 268 LEKSLGLDPNDPIIPFVV 285
+++ ++ I+PF V
Sbjct: 273 IKRVESIEKGGAIVPFEV 220
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.333 0.143 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,286,665
Number of Sequences: 36976
Number of extensions: 149589
Number of successful extensions: 1122
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 1116
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1121
length of query: 494
length of database: 9,014,727
effective HSP length: 100
effective length of query: 394
effective length of database: 5,317,127
effective search space: 2094948038
effective search space used: 2094948038
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.5 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146683.17


