

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146683.16 + phase: 0
(1672 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
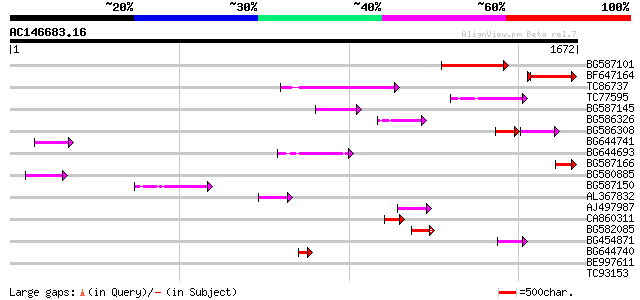
Score E
Sequences producing significant alignments: (bits) Value
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 309 5e-84
BF647164 weakly similar to GP|6691191|gb F7F22.15 {Arabidopsis t... 180 6e-51
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 189 8e-48
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 105 1e-22
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 93 8e-19
BG586326 similar to PIR|G84493|G8 probable retroelement pol poly... 87 7e-17
BG586308 weakly similar to PIR|F84528|F8 probable retroelement p... 59 9e-16
BG644741 79 2e-14
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 77 6e-14
BG587166 similar to GP|8778801|gb| T32E20.9 {Arabidopsis thalian... 67 6e-11
BG580885 65 2e-10
BG587150 similar to GP|13592175|gb ppg3 {Leishmania major}, part... 63 1e-09
AL367832 weakly similar to GP|14091845|gb Putative retroelement ... 62 2e-09
AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis... 58 3e-08
CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product ... 53 1e-06
BG582085 51 3e-06
BG454871 weakly similar to GP|10140673|g putative gag-pol polypr... 50 1e-05
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 46 1e-04
BE997611 39 0.013
TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Ci... 37 0.084
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 309 bits (792), Expect = 5e-84
Identities = 140/197 (71%), Positives = 165/197 (83%)
Frame = +2
Query: 1274 DLKHYYWDEPYLFRRGSDGIFRRCIPENEVSSILTHCHSSSYGGHASTQKTSFKILHSGF 1333
D++ Y WDEP+L+++ +D I+ RC+ E E+ IL HCH S+Y GH + KT KI +GF
Sbjct: 8 DVRRYLWDEPFLYKQCADNIYIRCVAEEEIPGILFHCHGSNYAGHFAVSKTVSKIQQAGF 187
Query: 1334 WWPSLFKDVHLFISKCDKCQRTGSITKRNEMPLNNILEVEIFDVWGIDFMGPFPSSFGNQ 1393
WWP++FKD H FISKCD CQR G+I+ RNEMP N ILEVE+FDVWGIDFMGPFPSS+ N+
Sbjct: 188 WWPTMFKDAHSFISKCDPCQRQGNIS*RNEMPQNFILEVEVFDVWGIDFMGPFPSSYNNK 367
Query: 1394 YILVAVDYVSKWVEAIASPTNDAQVVIKMFKKVIFPRFGVPRVVISDGGSHFISRHFEKL 1453
YILVAVDYVSKWVEAIASPTNDA VV+KMFK VIFPRFGVPRVVISDGGSHFI++ FEKL
Sbjct: 368 YILVAVDYVSKWVEAIASPTNDATVVVKMFKSVIFPRFGVPRVVISDGGSHFINKVFEKL 547
Query: 1454 LQKLGVRHKIATPYHPQ 1470
L+K GVRHK+AT YHPQ
Sbjct: 548 LKKNGVRHKVATAYHPQ 598
>BF647164 weakly similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana},
partial (6%)
Length = 469
Score = 180 bits (457), Expect(2) = 6e-51
Identities = 88/137 (64%), Positives = 106/137 (77%)
Frame = +1
Query: 1535 LEHKAYWAIRNLNLDPNLAGDKRKLQLNELEELRMDAYENARIYKERTKTWHDKKIIKRH 1594
L K I+ LN+D AG KR L+LNELEELR+DAYENA+IYKERTK WHD++II+R
Sbjct: 28 LSIKPIGPIKALNMDYKAAGGKRVLELNELEELRLDAYENAKIYKERTKKWHDRRIIRRE 207
Query: 1595 FKSGDLVLLFNSRLKLFPGKLRSRWSGPFQVRTVYPYGAIEIFSEETGSFTVNGQRLKIY 1654
F+ G+LVLLFNSRLKLFPGKLRS WSGPFQV+ V P GA+E++SE TG FTVNGQRLK Y
Sbjct: 208 FREGELVLLFNSRLKLFPGKLRSHWSGPFQVKNVMPSGAVEVWSESTGPFTVNGQRLKHY 387
Query: 1655 NTGEVNEVVADFTLSDP 1671
+GE + + L+ P
Sbjct: 388 TSGENVDKGVRYALTKP 438
Score = 40.8 bits (94), Expect(2) = 6e-51
Identities = 16/16 (100%), Positives = 16/16 (100%)
Frame = +3
Query: 1527 KSCHLPVELEHKAYWA 1542
KSCHLPVELEHKAYWA
Sbjct: 3 KSCHLPVELEHKAYWA 50
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 189 bits (480), Expect = 8e-48
Identities = 126/354 (35%), Positives = 177/354 (49%), Gaps = 3/354 (0%)
Frame = +1
Query: 798 VVKKEVLKLLDAGVIYPISDSKWVSPVQVVPKKGGLTVIKNDKNESIATRTVTGWRMCID 857
V+KK + LLD G I S S +PV V K GG G R C+D
Sbjct: 508 VLKKTLEDLLDKGFI-KASGSAAGAPVLFVRKPGG------------------GIRFCVD 630
Query: 858 YRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFTCPFG 917
YR LN T+KD +PLP I + L R+A F LD + F ++ I DQEKT F +G
Sbjct: 631 YRALNAITKKDRYPLPLISETLRRVAGARWFTKLDVVAAFHKMRIKDEDQEKTAFRTRYG 810
Query: 918 TFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGS-NFDDCLTNLEKVL 976
F + PFGL APATFQR + +F++ + ++DD ++ + + D + +VL
Sbjct: 811 LFEWIVCPFGLTGAPATFQRYINKTLHEFLDDFVTAYIDDVLIYTTGSKKDHEAQVRRVL 990
Query: 977 ERCEQVNLVLNWEKCHFMVREGIVLGH-LVFDRGIEVDRAKIEIIKKMLPPTSVKEIRSF 1035
R L L+ +KC F V +G L +G+ D K+ I+ LPP SVK RSF
Sbjct: 991 RRLADAGLSLDPKKCEFSVTTVKYVGFILTAGKGVSCDPLKLAAIRDWLPPGSVKGARSF 1170
Query: 1036 LGHAGFYRRFIKDFSSITKPLTSLLLKDADFTFDDSCLQAFCRLKEALITAPIIQPPDWN 1095
LG +Y+ FI +S IT+PLT L KD F + AF +LK P+++ D
Sbjct: 1171 LGFCNYYKDFIPGYSEITEPLTRLTRKDFPFRWGAEQEAAFTKLKRLFAEEPVLRMFDPE 1350
Query: 1096 LPFEIMCDASDYAVGAVLGQRNDK-KMHAIYYASKTLDGAQVNYATTEKELLAV 1148
+ D S +A+G VL Q + H + + S+ L A+ NY +KELLAV
Sbjct: 1351 AVTTVETDCSGFALGGVLTQEDGTGAAHPVAFHSQRLSPAEYNYPIHDKELLAV 1512
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial (14%)
Length = 1708
Score = 105 bits (263), Expect = 1e-22
Identities = 69/236 (29%), Positives = 116/236 (48%), Gaps = 8/236 (3%)
Frame = +2
Query: 1299 PENEV-SSILTHCHSSSYGGHASTQKTSFKILHSGFWWPSLFKDVHLFISKCDKCQ---- 1353
P NE+ + ++ H S+ GH T +I+ F+WP + V F+ CD C
Sbjct: 155 PLNELRTKLVQESHDSTAAGHPGRNGT-LEIVSRKFFWPGQSQTVRRFVRNCDVCGGIHI 331
Query: 1354 -RTGSITKRNEMPLNNILEVEIFDVWGIDFMGPFPSSFG--NQYILVAVDYVSKWVEAIA 1410
R +P+ N L ++ +DF+ P + G +QY+ V VD +SK V
Sbjct: 332 WRQAKRGFLKPLPVPNRLHSDL----SMDFITSLPPTRGRGSQYLWVIVDRLSKSVTLEE 499
Query: 1411 SPTNDAQVVIKMFKKVIFPRFGVPRVVISDGGSHFISRHFEKLLQKLGVRHKIATPYHPQ 1470
T +A+ + F + G+P+ ++SD GS+++ R + + + GV ++T YHPQ
Sbjct: 500 MDTMEAEACAQRFLSCHYRFHGMPQSIVSDRGSNWVGRFWREFCRLTGVTQLLSTSYHPQ 679
Query: 1471 TSGQVEVSNRQIKAILEKTVSTSRTDWSNKLDDALWAYRTAYKTPIGMTPFKLVYG 1526
T G E N++I+A+L V S+ +W + L A R + + IG TPF + +G
Sbjct: 680 TDGGTERWNQEIQAVLRAYVCWSQDNWGDLLPTVQLALRNRHNSSIGATPFFVEHG 847
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 93.2 bits (230), Expect = 8e-19
Identities = 50/136 (36%), Positives = 79/136 (57%)
Frame = +2
Query: 902 IHPNDQEKTTFTCPFGTFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVH 961
+HP+D EKT F GT+ Y+ MPFGL NA +T+QR + +F+D + MEV++DD V
Sbjct: 2 MHPDDLEKTAFITDRGTYCYKVMPFGLKNAGSTYQRLVNRMFADKLGNTMEVYIDDMLVK 181
Query: 962 GSNFDDCLTNLEKVLERCEQVNLVLNWEKCHFMVREGIVLGHLVFDRGIEVDRAKIEIIK 1021
D L +L++ + ++ + LN KC F V G LG++V +GIEV+ +I I
Sbjct: 182 SLRATDHLNHLKE*FKTLDEYIMKLNPAKCTFGVTSGEFLGYIVTQQGIEVNPKQITAIL 361
Query: 1022 KMLPPTSVKEIRSFLG 1037
+ P + +E++ G
Sbjct: 362 DLPSPKNSREVQRLTG 409
>BG586326 similar to PIR|G84493|G8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 736
Score = 86.7 bits (213), Expect = 7e-17
Identities = 52/145 (35%), Positives = 81/145 (55%)
Frame = +2
Query: 1085 TAPIIQPPDWNLPFEIMCDASDYAVGAVLGQRNDKKMHAIYYASKTLDGAQVNYATTEKE 1144
+API+ P+ + + + DAS +G VL Q I YAS+ L + NY T + E
Sbjct: 11 SAPILVLPEL-ITYVVYTDASITGLGCVLTQHEK----VIAYASRQLRKHEGNYPTHDLE 175
Query: 1145 LLAVVYAIDKFRQYLVGSKIIVYTDHSAIKYLLNKKDAKPRLIRWILLLQEFDLEIKDKK 1204
+ AVV+A+ +R YL G+K+ ++TDH ++KY+ + + R RW+ + ++DL+I
Sbjct: 176 MAAVVFALKIWRSYLYGAKVQIHTDHKSLKYIFTQPELNLRQRRWMEFVADYDLDITYYP 355
Query: 1205 GVENVVADHLSRLRETNKDELPLDD 1229
G N+VAD LSR R E DD
Sbjct: 356 GKANLVADALSRRRVDVSAEREADD 430
>BG586308 weakly similar to PIR|F84528|F8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 686
Score = 58.5 bits (140), Expect(2) = 9e-16
Identities = 26/71 (36%), Positives = 45/71 (62%)
Frame = -2
Query: 1432 GVPRVVISDGGSHFISRHFEKLLQKLGVRHKIATPYHPQTSGQVEVSNRQIKAILEKTVS 1491
G+P +++D GSHFIS F + ++ +R A+P +PQ++GQ E SN+ I L+K +
Sbjct: 682 GLPYEIVTDNGSHFISNKFREFCERWRIRLNTASPRYPQSNGQAEASNKIIIDGLKKRLD 503
Query: 1492 TSRTDWSNKLD 1502
+ W+++LD
Sbjct: 502 LKKGCWADELD 470
Score = 44.7 bits (104), Expect(2) = 9e-16
Identities = 31/119 (26%), Positives = 57/119 (47%), Gaps = 2/119 (1%)
Frame = -3
Query: 1505 LWAYRTAYKTPIGMTPFKLVYGKSCHLPVELEHKAYWAIRNLNLDPNLAGDKRKLQLNEL 1564
LW++RT + TPF + + P E+ + R + + L D+ L +
Sbjct: 462 LWSHRTNPRGATKSTPFSMAHRVEAMAPAEVNVTSL*RSR-MPQNIELNNDRLFNALETI 286
Query: 1565 EELRMDAYENARIYKERTKTWHDKKIIKRHFKSGDLVL--LFNSRLKLFPGKLRSRWSG 1621
EE R A + Y+ + +++++K + + K GD+VL +F + +L GKL + W G
Sbjct: 285 EERRDQALLRIQNYQHQIESYYNKTVKSQPLKLGDIVLCKVFENTKELNAGKLGTNWEG 109
>BG644741
Length = 735
Score = 78.6 bits (192), Expect = 2e-14
Identities = 40/115 (34%), Positives = 59/115 (50%)
Frame = -2
Query: 72 DQETVRLYLFPFSLKDKASNWFNSLKPGSITSWDQLRREFLCRFFPPSKTAQLRGKLYQF 131
D + L +FP SL +A WF L SI +W+QLR FL R++P SK ++ F
Sbjct: 605 DLNVLGLRVFPLSLMGEADIWFTELPYNSIFTWNQLRDVFLARYYPVSKKLNHNDRVNNF 426
Query: 132 TQKNEESLFDVWEHFKEILRRCPHHGLEKWLIIHTFYNGLTSSTKLTVDTAAGGA 186
ES+ W+ F LR P+H ++ + FY G + K+ +DT AGG+
Sbjct: 425 VALPGESVSSSWDRFTSFLRSVPNHRIDDDSLKEYFYRGQDDNNKVVLDTIAGGS 261
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 77.0 bits (188), Expect = 6e-14
Identities = 59/224 (26%), Positives = 98/224 (43%)
Frame = +2
Query: 790 RLNPNMKEVVKKEVLKLLDAGVIYPISDSKWVSPVQVVPKKGGLTVIKNDKNESIATRTV 849
R+NP +V+K ++ LL+ G I P + P G+ V+ K +
Sbjct: 92 RINPLKLKVLKLQLKDLLEKGFIQP----------SIYP*--GVVVLFLKKKDGFL---- 223
Query: 850 TGWRMCIDYRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEK 909
RM IDY +LN K +PLP ID++ + L F +D G Q + D K
Sbjct: 224 ---RMSIDYPQLNNVNIKIKYPLPLIDELFDNLQGSKWFFKIDLRLG*HQHRVIGEDVPK 394
Query: 910 TTFTCPFGTFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCL 969
T F +G + M FG N P F M +F D+++ ++ VF +D ++ N ++
Sbjct: 395 TAFRIRYGHYEILVMSFG*TNPPMAFMELMNRVFQDYLDSLVIVFSNDILIYSKNENEHE 574
Query: 970 TNLEKVLERCEQVNLVLNWEKCHFMVREGIVLGHLVFDRGIEVD 1013
+L L+ + + L +V G H++ G++VD
Sbjct: 575 NHLRLALKVLKDIGL-CQISYV*ILVEVGFFSLHVISGEGLKVD 703
>BG587166 similar to GP|8778801|gb| T32E20.9 {Arabidopsis thaliana}, partial
(2%)
Length = 756
Score = 67.0 bits (162), Expect = 6e-11
Identities = 34/62 (54%), Positives = 42/62 (66%)
Frame = +1
Query: 1610 LFPGKLRSRWSGPFQVRTVYPYGAIEIFSEETGSFTVNGQRLKIYNTGEVNEVVADFTLS 1669
LFPGKL+SR SGPF+V+ V PYGAI ++S + FTVNGQR+K+Y E LS
Sbjct: 1 LFPGKLKSR*SGPFKVKEVRPYGAIVLWSTDGRDFTVNGQRVKLYMATAPEEDGVSTPLS 180
Query: 1670 DP 1671
DP
Sbjct: 181 DP 186
>BG580885
Length = 609
Score = 65.5 bits (158), Expect = 2e-10
Identities = 37/127 (29%), Positives = 70/127 (54%), Gaps = 2/127 (1%)
Frame = +3
Query: 47 FSGSPSEDPNLHISTFWRLSVTCKDDQETVRLYLFPFSLKDKASNWFN-SLKPGSIT-SW 104
F G+P+E P H+S F ++ ++ +FP +L+++++ W++ +++P I+ SW
Sbjct: 54 FRGTPNESPITHLSRFNKVCRANNASSVEMQKKIFPVTLEEESALWYDLNIEPYYISLSW 233
Query: 105 DQLRREFLCRFFPPSKTAQLRGKLYQFTQKNEESLFDVWEHFKEILRRCPHHGLEKWLII 164
D+++ FL ++ +LR +L Q +E + + + IL+R P HGLE +I
Sbjct: 234 DEIKLSFLQAYYEIEPVEELRSELMGIHQGEKERVRSYFLRLQWILKRWPEHGLEDDVIK 413
Query: 165 HTFYNGL 171
F NGL
Sbjct: 414 GVFVNGL 434
>BG587150 similar to GP|13592175|gb ppg3 {Leishmania major}, partial (1%)
Length = 754
Score = 62.8 bits (151), Expect = 1e-09
Identities = 57/230 (24%), Positives = 100/230 (42%)
Frame = +2
Query: 367 KPLEDPIRRTETDNSEKEICEPPSRETRAEREKPRVEENLPPPFKPKIPFPQRFAKSKLD 426
KP ++P T T + + E ++ + NL K +R A K
Sbjct: 5 KPSDNPPEATSTHSDDAA--------GPMEVDRVHMGRNLRKRKKKVAKHLKRGANEKEM 160
Query: 427 EQFKKFIEMMNKIYIDVPFTEVLTQMPTYAKFLKEILSKKRKIEESETVNLTEECSAIIQ 486
E F+K + M I ++ PF E + L + R+ EE E + +
Sbjct: 161 ESFQKRVFM---IPLEKPFEEAY-----FTHRLWMFFRETRETEEDIRRMFCEAREKMKK 316
Query: 487 NKLPPKLKDPGSFSIPCVIGSEVVKKAMCDLGASVSLMPLSLYERLGIGELKSTRMTLQL 546
K D G F+I + A+C+ GASV ++P + + LG+ ++ ++ +
Sbjct: 317 RITLKKKSDSGKFAISWTMKGIEFPHALCNTGASVIILPRDMADHLGL-KVDPSKESFTF 493
Query: 547 ADRSVKYPAGIIEDVPVKVGEVYIPADFVVMEMEEDNQVPILLGRPFLAT 596
D S + GII D+ V++G +P DF V++++ + +LL R FL+T
Sbjct: 494 VDCSQRSSGGIIRDLEVQIGNALVPVDFHVLDIKINWNSSLLLRRAFLST 643
>AL367832 weakly similar to GP|14091845|gb Putative retroelement {Oryza
sativa}, partial (2%)
Length = 384
Score = 62.0 bits (149), Expect = 2e-09
Identities = 32/100 (32%), Positives = 55/100 (55%)
Frame = -1
Query: 733 VNASLDEVETEKLLYVLKKYPKAIGYTIDDIKGINPSLCMHRILLEEDYKPSIEHQRRLN 792
V A+L+E K+ +L++Y + +D+ G++P + HRI + + P RR +
Sbjct: 300 VGAALEEGVKRKIFQLLREYLDIFACSYEDMPGLDPKIVEHRIPTKPECPPVR*KLRRTH 121
Query: 793 PNMKEVVKKEVLKLLDAGVIYPISDSKWVSPVQVVPKKGG 832
P+M +K EV K +DAG + + +WV+ + VPKK G
Sbjct: 120 PDMALKIKSEVQKQIDAGFLMTVEYPEWVANIVPVPKKDG 1
>AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis thaliana
chromosome II BAC F26H6; putative retroelement pol
polyprotein, partial (1%)
Length = 636
Score = 58.2 bits (139), Expect = 3e-08
Identities = 29/102 (28%), Positives = 55/102 (53%), Gaps = 1/102 (0%)
Frame = -2
Query: 1143 KELLAVVYAIDKFRQYLVGSKIIVYTDHSAIKYLLNKKDAKPRLIRWILLLQEFDLEIKD 1202
K A+ +A + R Y++ + + IKY+ K R+ RW +LL E+D+E +
Sbjct: 635 KTCCALAWAAKRLRHYMINHTTWLVSKMDPIKYIFEKPALTGRIARWQMLLSEYDIEYRS 456
Query: 1203 KKGVE-NVVADHLSRLRETNKDELPLDDSFPDDQLFLLAQTD 1243
+K ++ +++ADHL+ + +D P+ FPD+++ L D
Sbjct: 455 QKAIKGSILADHLA--HQPLEDYRPIKFDFPDEEIMYLKMKD 336
>CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product {Drosophila
melanogaster}, partial (20%)
Length = 192
Score = 52.8 bits (125), Expect = 1e-06
Identities = 27/61 (44%), Positives = 38/61 (62%), Gaps = 1/61 (1%)
Frame = +1
Query: 1104 ASDYAVGAVLGQRNDKKM-HAIYYASKTLDGAQVNYATTEKELLAVVYAIDKFRQYLVGS 1162
A D+ +GA L Q++++ H I YAS+ L A+ NY E+E LA ++AI FR YL G
Sbjct: 1 APDWGIGAGLSQKDEENHEHPIAYASRLLTAAERNYTVVERECLAAIWAIRNFRHYLHGP 180
Query: 1163 K 1163
K
Sbjct: 181 K 183
>BG582085
Length = 824
Score = 51.2 bits (121), Expect = 3e-06
Identities = 27/72 (37%), Positives = 46/72 (63%), Gaps = 2/72 (2%)
Frame = -3
Query: 1184 PRLIRWILLLQEFDLEIKDKK--GVENVVADHLSRLRETNKDELPLDDSFPDDQLFLLAQ 1241
P +R ILL QEFDLE + ++ G+ ++A R ++ ++E+ + + FP++QLF ++Q
Sbjct: 771 P*RLRCILLCQEFDLESRQERETGMW*LIAFPCLRNKQVTQNEISIKERFPNEQLFAISQ 592
Query: 1242 TDAPWYADFVNF 1253
PW+AD NF
Sbjct: 591 --RPWFADMTNF 562
>BG454871 weakly similar to GP|10140673|g putative gag-pol polyprotein {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 674
Score = 49.7 bits (117), Expect = 1e-05
Identities = 27/88 (30%), Positives = 43/88 (48%)
Frame = +2
Query: 1440 DGGSHFISRHFEKLLQKLGVRHKIATPYHPQTSGQVEVSNRQIKAILEKTVSTSRTDWSN 1499
+G S S +++L + G +++ YHP + GQ E N+ + L + T WS
Sbjct: 11 EG*SSLYSNFWKQLFKLHGTILTMSSAYHP*SDGQSEALNKGXEMYLRCLMFTDPLKWSK 190
Query: 1500 KLDDALWAYRTAYKTPIGMTPFKLVYGK 1527
A + Y T+Y MTPFK +YG+
Sbjct: 191 AFPWAEYWYNTSYNISAAMTPFKALYGR 274
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 45.8 bits (107), Expect = 1e-04
Identities = 18/41 (43%), Positives = 29/41 (69%)
Frame = -1
Query: 852 WRMCIDYRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLD 892
+RMCIDYR+ NK T K+ +PLP ID + +++ + +F +D
Sbjct: 202 FRMCIDYRQFNKVTTKNKYPLPRIDNLFDKIQEDCYF*NID 80
>BE997611
Length = 547
Score = 39.3 bits (90), Expect = 0.013
Identities = 20/92 (21%), Positives = 45/92 (48%), Gaps = 3/92 (3%)
Frame = -2
Query: 422 KSKLDEQFKKFIEMMNKIYIDVPFTEVLTQMPTYAKFLKEILSKKRKIEESETVNLTEEC 481
+ ++ +F ++ +++P TE + Q+P KFL+E+L + E E ++L+ E
Sbjct: 444 EKEVPPKFNALHHILKGYKVNMPITETVEQIPCCIKFLQELLKTNANLSEEEFISLSSEF 265
Query: 482 SAIIQNKLPPKLKDPGSFS---IPCVIGSEVV 510
+ + G F+ +P +IG+ ++
Sbjct: 264 HHTYEVPAVVRFDGEGCFTLRFVPRIIGTHLI 169
>TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Cicer
arietinum}, partial (8%)
Length = 516
Score = 36.6 bits (83), Expect = 0.084
Identities = 23/91 (25%), Positives = 42/91 (45%), Gaps = 5/91 (5%)
Frame = +2
Query: 74 ETVRLYLFPFSLKDKASNWFNSLKP-----GSITSWDQLRREFLCRFFPPSKTAQLRGKL 128
ET ++ L ++A +W+ SL P ++ +W R+EFL R+FP + +
Sbjct: 221 ETQKVQFGTHMLAEEADDWWISLLPVLEQDDAVVTWAMFRKEFLGRYFPEDVRGKKEIEF 400
Query: 129 YQFTQKNEESLFDVWEHFKEILRRCPHHGLE 159
+ Q + S+ + F E+ PH+ E
Sbjct: 401 LELKQ-GDMSVTEYAAKFVELATFYPHYSAE 490
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.137 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 52,905,328
Number of Sequences: 36976
Number of extensions: 784379
Number of successful extensions: 3793
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 3724
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3779
length of query: 1672
length of database: 9,014,727
effective HSP length: 109
effective length of query: 1563
effective length of database: 4,984,343
effective search space: 7790528109
effective search space used: 7790528109
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 65 (29.6 bits)
Medicago: description of AC146683.16


