

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146664.12 + phase: 0 /pseudo
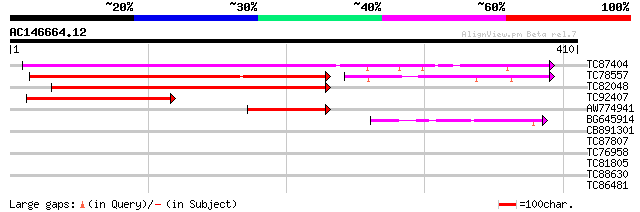
(410 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC87404 similar to GP|22137130|gb|AAM91410.1 At1g47330/T3F24_2 {... 285 2e-77
TC78557 similar to PIR|A71404|A71404 hypothetical protein - Arab... 211 2e-67
TC82048 similar to GP|12039276|gb|AAG46066.1 putative ancient co... 236 1e-62
TC92407 similar to PIR|B84518|B84518 hypothetical protein At2g14... 97 9e-21
AW774941 similar to GP|17381276|gb| AT4g33700/T16L1_190 {Arabido... 94 1e-19
BG645914 similar to PIR|A71404|A71 hypothetical protein - Arabid... 59 4e-09
CB891301 homologue to PIR|A71404|A714 hypothetical protein - Ara... 39 0.004
TC87807 similar to GP|6175161|gb|AAF04887.1| unknown protein {Ar... 29 2.9
TC76958 homologue to SP|P41382|IF4U_TOBAC Eukaryotic initiation ... 29 3.8
TC81805 similar to GP|9758323|dbj|BAB08797.1 Myb-related transcr... 28 5.0
TC88630 similar to GP|22655068|gb|AAM98125.1 unknown protein {Ar... 28 8.5
TC86481 similar to GP|9294170|dbj|BAB02072.1 emb|CAB71043.1~gene... 28 8.5
>TC87404 similar to GP|22137130|gb|AAM91410.1 At1g47330/T3F24_2 {Arabidopsis
thaliana}, partial (70%)
Length = 1944
Score = 285 bits (729), Expect = 2e-77
Identities = 173/435 (39%), Positives = 251/435 (56%), Gaps = 50/435 (11%)
Frame = +3
Query: 10 DEHCCGNHFWVLALLCWVFMFFAAISSALALGLLSFSQVDLEVLVKAGQPHIQKNAAKIM 69
D CCG FW+ L+ + FA + + L LGL+S VDLEVL+K+G+P + +AAKI
Sbjct: 108 DVGCCGTKFWLYILMIIGLVCFAGLMAGLTLGLMSLGLVDLEVLIKSGRPQHRIHAAKIF 287
Query: 70 SIVKNEHLVLCTLLMAKSLALEGVSVLMEKMFPEWVAVLLATALISIIAEVIPQALNSRY 129
+VKN+HL+LCTLL+ SLA+E + + ++ + P + AVL++ LI I E++PQA+ +RY
Sbjct: 288 PVVKNQHLLLCTLLIGNSLAMETLPIFLDAIVPPYAAVLISVTLILIFGEILPQAVCTRY 467
Query: 130 GLRFGATMSPFVRVLLLLFFPFAYPVSKLLDCLLGKGHTALLGREELKTLVNLHANEAGK 189
GL GAT++P VRVLLL+F+P AYP+SK+LD +LGKG ALL R ELKT V+ H NEAGK
Sbjct: 468 GLLVGATLAPLVRVLLLVFYPIAYPISKVLDRMLGKGKAALLKRAELKTFVDFHGNEAGK 647
Query: 190 GGELTLHETTIIAGALDLTMKTAKDAMTPLSETFSLDINSKLDMSRI*CSAVRKMKHQLS 249
GG+LT ETTIIAGAL+LT KTAKDAMTP+S+ FSLD+++ L++ + +++ M H
Sbjct: 648 GGDLTHDETTIIAGALELTEKTAKDAMTPISKAFSLDLDATLNLETL--NSIMTMGHSRV 821
Query: 250 L*L*GEFP---------------------------------GENWPLYDILNQFKKGQSH 276
GE EN PLYDILN+F+KG SH
Sbjct: 822 PVYAGERTNIMGLVLVKNLFMVDSKAAVPLRKMIIRKIPRVSENMPLYDILNEFQKGHSH 1001
Query: 277 MAVV---LKSKENIRTAATNTEGF---------GPFLPHDYISISTEASNWQSEGSEYYS 324
+AVV L K+ I N E G P D ++ ++ + + GS+
Sbjct: 1002IAVVYRDLNDKKEISKKIKNEEQLEFKDSCRNKGKSAPLDKGTV-LDSHDTVTAGSK--- 1169
Query: 325 ATLKNAMLQESKDSDPLHRSKQHDTSISLENMES-----LLGEEEVVGIITLEDVMEELL 379
T +++S + P + + S + ++++ E VVG+I++EDV+EELL
Sbjct: 1170-TDGGPQVKKSPPATPAFKKRHRGCSYCILDLDNAPLPVFPPNEVVVGVISMEDVIEELL 1346
Query: 380 QVSLTSYHSTIIFVH 394
Q + + +H
Sbjct: 1347QEEILDETDEYVNIH 1391
>TC78557 similar to PIR|A71404|A71404 hypothetical protein - Arabidopsis
thaliana, partial (72%)
Length = 1690
Score = 211 bits (538), Expect(2) = 2e-67
Identities = 108/219 (49%), Positives = 155/219 (70%), Gaps = 1/219 (0%)
Frame = +2
Query: 15 GNHFWVLAL-LCWVFMFFAAISSALALGLLSFSQVDLEVLVKAGQPHIQKNAAKIMSIVK 73
G+ W+ + +C + FA I S L LGL+S S VDLE+L ++G P +K AA I+ +VK
Sbjct: 164 GSVTWIAYVGICCFLVCFAGIMSGLTLGLMSLSLVDLEILERSGSPSEKKQAAIILPVVK 343
Query: 74 NEHLVLCTLLMAKSLALEGVSVLMEKMFPEWVAVLLATALISIIAEVIPQALNSRYGLRF 133
+H +L TLL+ ++A+E + + ++K+F +++A++L+ + EVIPQA+ SRYGL
Sbjct: 344 KQHQLLVTLLLCNAVAMEALPIYLDKLFNQFLAIILSVTFVLFFGEVIPQAICSRYGLAV 523
Query: 134 GATMSPFVRVLLLLFFPFAYPVSKLLDCLLGKGHTALLGREELKTLVNLHANEAGKGGEL 193
GA + VR+L+++ +P +YPV K+LD LLG AL R +LK LV++H EAGKGGEL
Sbjct: 524 GANFAWLVRILMVICYPVSYPVGKVLDYLLGHDE-ALFRRAQLKALVSIHGKEAGKGGEL 700
Query: 194 TLHETTIIAGALDLTMKTAKDAMTPLSETFSLDINSKLD 232
T ETTII+GALDLT KTA++AMTP+ TFSLD+NSKLD
Sbjct: 701 THDETTIISGALDLTEKTAEEAMTPIESTFSLDVNSKLD 817
Score = 62.8 bits (151), Expect(2) = 2e-67
Identities = 52/164 (31%), Positives = 75/164 (45%), Gaps = 12/164 (7%)
Frame = +1
Query: 243 KMKHQLSL*L*GEFPG--ENWPLYDILNQFKKGQSHMAVVLKSKENIRTAATNTEGFGPF 300
K++ L+L L GEFP + PLYDILN+F+KG SHMA V+K+K G G
Sbjct: 937 KLRPLLALYLSGEFPRVPSDMPLYDILNEFQKGSSHMAAVVKAK-----------GKGKE 1083
Query: 301 LPHDYISISTEASNWQSEGSEYYSATLKNAMLQESK---DSDPLHRSKQHDTSISLENME 357
P +A S+ + L+ ++ D D + + L+ +
Sbjct: 1084 TPQIIDEEKFDAKKSAGGESQLTTPLLQKQDVKSENVVVDIDEPSKLPIINKQTGLQRSD 1263
Query: 358 SLLG-------EEEVVGIITLEDVMEELLQVSLTSYHSTIIFVH 394
+L + EV+GIITLEDV EELLQ + + VH
Sbjct: 1264 GMLNTPSENFEDGEVIGIITLEDVFEELLQEEIVDETDEYVDVH 1395
>TC82048 similar to GP|12039276|gb|AAG46066.1 putative ancient conserved
domain protein {Oryza sativa}, partial (78%)
Length = 856
Score = 236 bits (602), Expect = 1e-62
Identities = 116/202 (57%), Positives = 157/202 (77%)
Frame = +2
Query: 31 FAAISSALALGLLSFSQVDLEVLVKAGQPHIQKNAAKIMSIVKNEHLVLCTLLMAKSLAL 90
FA + S L LGL+S S VDLEVL K+G P+ +K+A KI+ +V+N+HL+LCTLL+ + A+
Sbjct: 179 FAGLMSGLTLGLMSLSLVDLEVLAKSGTPNDRKHAVKILPVVRNQHLLLCTLLICNAAAM 358
Query: 91 EGVSVLMEKMFPEWVAVLLATALISIIAEVIPQALNSRYGLRFGATMSPFVRVLLLLFFP 150
E + + ++ + W A+L++ LI + E+IPQ++ SRYGL GA+++PFVRVL+ + +P
Sbjct: 359 EALPIFLDSLVTAWGAILISVTLILLFGEIIPQSVCSRYGLAIGASVTPFVRVLVWICYP 538
Query: 151 FAYPVSKLLDCLLGKGHTALLGREELKTLVNLHANEAGKGGELTLHETTIIAGALDLTMK 210
A+P+SKLLD LLG + AL R ELKTLV+LH NEAGKGGELT ETTIIAGAL+L+ K
Sbjct: 539 VAFPISKLLDYLLGHRNEALFRRAELKTLVDLHGNEAGKGGELTHDETTIIAGALELSEK 718
Query: 211 TAKDAMTPLSETFSLDINSKLD 232
TA DAMTP+SETF++DINSKLD
Sbjct: 719 TASDAMTPISETFAIDINSKLD 784
>TC92407 similar to PIR|B84518|B84518 hypothetical protein At2g14520
[imported] - Arabidopsis thaliana, partial (27%)
Length = 619
Score = 97.4 bits (241), Expect = 9e-21
Identities = 43/108 (39%), Positives = 71/108 (64%)
Frame = +2
Query: 13 CCGNHFWVLALLCWVFMFFAAISSALALGLLSFSQVDLEVLVKAGQPHIQKNAAKIMSIV 72
CC F++ ++ + + FA + S L LGL+S S VDLEVL K+G P +K+A KI+ +V
Sbjct: 296 CCETEFFIRIMIIVLLVVFAGLMSGLTLGLMSLSLVDLEVLAKSGTPQDRKHAEKILPVV 475
Query: 73 KNEHLVLCTLLMAKSLALEGVSVLMEKMFPEWVAVLLATALISIIAEV 120
KN+HL+LCTLL+ + A+E + + ++ + W A+L++ LI + E+
Sbjct: 476 KNQHLLLCTLLICNAAAMEALPIFLDSLVVAWGAILISVTLILLFGEI 619
>AW774941 similar to GP|17381276|gb| AT4g33700/T16L1_190 {Arabidopsis
thaliana}, partial (16%)
Length = 215
Score = 93.6 bits (231), Expect = 1e-19
Identities = 47/60 (78%), Positives = 53/60 (88%)
Frame = +1
Query: 173 REELKTLVNLHANEAGKGGELTLHETTIIAGALDLTMKTAKDAMTPLSETFSLDINSKLD 232
R ELKTLVNLH NEAGKGGELT ETTIIAGAL+L+ KTA DAMTP++E FS+DINSKL+
Sbjct: 4 RAELKTLVNLHGNEAGKGGELTHDETTIIAGALELSEKTAGDAMTPINEIFSIDINSKLN 183
>BG645914 similar to PIR|A71404|A71 hypothetical protein - Arabidopsis
thaliana, partial (19%)
Length = 672
Score = 58.5 bits (140), Expect = 4e-09
Identities = 50/135 (37%), Positives = 65/135 (48%), Gaps = 7/135 (5%)
Frame = -1
Query: 262 PLYDILNQFKKGQSHMAVVLKSKENIRTAATNTEGFGPFLPHDYISISTEASNWQSEGSE 321
PLYDILN+F+KG SHMA V+K T G G P + + ++ S
Sbjct: 588 PLYDILNEFQKGSSHMAAVVK-----------TRGKGKETPQ-----LIDEEKFDAKKSV 457
Query: 322 YYSATLKNAMLQESKDSDPLHRSKQHDTSISLENMESLLGEEEVVGIITLEDVMEE---- 377
+ L +LQE DS + D L + S + EV+GIITLEDV EE
Sbjct: 456 GGDSQLTTPLLQE-LDSKSENVVVDIDKPSKLTSAPSENIDGEVIGIITLEDVFEELLQV 280
Query: 378 ---LLQVSLTSYHST 389
LLQV+L+S H T
Sbjct: 279 NFCLLQVNLSSAHKT 235
>CB891301 homologue to PIR|A71404|A714 hypothetical protein - Arabidopsis
thaliana, partial (10%)
Length = 638
Score = 38.9 bits (89), Expect = 0.004
Identities = 16/23 (69%), Positives = 21/23 (90%)
Frame = +1
Query: 210 KTAKDAMTPLSETFSLDINSKLD 232
+TA++AMTP+ TFSLD+NSKLD
Sbjct: 478 QTAEEAMTPIESTFSLDVNSKLD 546
>TC87807 similar to GP|6175161|gb|AAF04887.1| unknown protein {Arabidopsis
thaliana}, partial (64%)
Length = 1398
Score = 29.3 bits (64), Expect = 2.9
Identities = 16/54 (29%), Positives = 29/54 (53%)
Frame = +1
Query: 311 EASNWQSEGSEYYSATLKNAMLQESKDSDPLHRSKQHDTSISLENMESLLGEEE 364
EA + +S S+Y+ A L+ AML +DP R+ H + +N + L +++
Sbjct: 376 EAGSVKSGVSDYFFAILEAAMLSPVSCTDPSVRADYHSLFTAAQNAKEHLNKKQ 537
>TC76958 homologue to SP|P41382|IF4U_TOBAC Eukaryotic initiation factor
4A-10 (eIF-4A-10) (eIF4A-10). [Common tobacco]
{Nicotiana tabacum}, complete
Length = 1761
Score = 28.9 bits (63), Expect = 3.8
Identities = 12/48 (25%), Positives = 24/48 (50%)
Frame = +1
Query: 300 FLPHDYISISTEASNWQSEGSEYYSATLKNAMLQESKDSDPLHRSKQH 347
F+ H + S+ +WQ+ +S L + + + + S P+HR+ H
Sbjct: 124 FISHRSSTFSSSLKSWQALHQRVHSLMLSSLIQK*TSCSQPMHRTSSH 267
>TC81805 similar to GP|9758323|dbj|BAB08797.1 Myb-related transcription
factor-like protein {Arabidopsis thaliana}, partial
(46%)
Length = 1221
Score = 28.5 bits (62), Expect = 5.0
Identities = 16/33 (48%), Positives = 21/33 (63%)
Frame = -1
Query: 134 GATMSPFVRVLLLLFFPFAYPVSKLLDCLLGKG 166
GAT+ + +LLLL FP A V++LL L KG
Sbjct: 777 GATLECMIELLLLLLFPEALKVTELL-LLFSKG 682
>TC88630 similar to GP|22655068|gb|AAM98125.1 unknown protein {Arabidopsis
thaliana}, partial (2%)
Length = 1128
Score = 27.7 bits (60), Expect = 8.5
Identities = 13/34 (38%), Positives = 19/34 (55%)
Frame = -3
Query: 14 CGNHFWVLALLCWVFMFFAAISSALALGLLSFSQ 47
C +FW C+V + F+ S L +G LSFS+
Sbjct: 916 CAGYFW-----CYVMLSFSRFSYHLFIGRLSFSR 830
>TC86481 similar to GP|9294170|dbj|BAB02072.1
emb|CAB71043.1~gene_id:MJL12.8~similar to unknown protein
{Arabidopsis thaliana}, partial (86%)
Length = 2040
Score = 27.7 bits (60), Expect = 8.5
Identities = 14/38 (36%), Positives = 20/38 (51%), Gaps = 6/38 (15%)
Frame = +2
Query: 11 EHCCGNHFWVLALLCWVFMFFAAI------SSALALGL 42
E C N FWV++ +C V F A++ S L +GL
Sbjct: 1346 EWSCRNMFWVIS*ICTVHEFLASVDQS*I*SEGLCMGL 1459
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.324 0.137 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,249,398
Number of Sequences: 36976
Number of extensions: 155941
Number of successful extensions: 1336
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 1328
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1333
length of query: 410
length of database: 9,014,727
effective HSP length: 98
effective length of query: 312
effective length of database: 5,391,079
effective search space: 1682016648
effective search space used: 1682016648
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146664.12


