

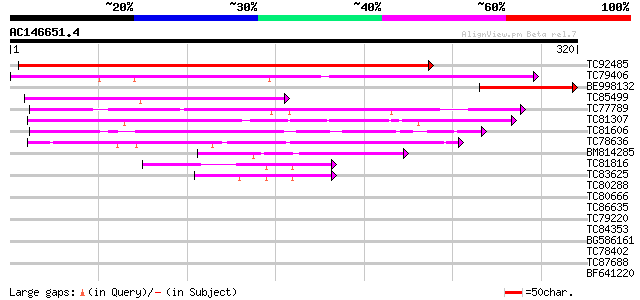
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146651.4 - phase: 0
(320 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC92485 similar to GP|21592663|gb|AAM64612.1 unknown {Arabidopsi... 430 e-121
TC79406 weakly similar to PIR|T04879|T04879 hypothetical protein... 196 8e-51
BE998132 homologue to GP|21592663|gb| unknown {Arabidopsis thali... 112 1e-25
TC85499 similar to GP|22477284|gb|AAH28621.1 Unknown (protein fo... 99 3e-21
TC77789 weakly similar to GP|6671937|gb|AAF23197.1| unknown prot... 78 4e-15
TC81307 weakly similar to GP|4335772|gb|AAD17449.1| unknown prot... 77 1e-14
TC81606 similar to EGAD|129691|138422 fibroblast growth factor 1... 67 9e-12
TC78636 45 3e-05
BM814285 weakly similar to GP|9295718|gb| T24P13.3 {Arabidopsis ... 45 3e-05
TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana taba... 43 1e-04
TC83625 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana taba... 42 4e-04
TC80288 weakly similar to PIR|T06029|T06029 hypothetical protein... 40 0.002
TC80666 similar to PIR|E85112|E85112 probable methyltransferase ... 35 0.039
TC86635 weakly similar to GP|9758600|dbj|BAB09233.1 gene_id:MDF2... 33 0.15
TC79220 similar to GP|2149570|gb|AAB58577.1| MAP kinase kinase p... 33 0.15
TC84353 similar to GP|20466332|gb|AAM20483.1 putative protein {A... 33 0.19
BG586161 similar to GP|21554129|gb| putative thaumatin {Arabidop... 32 0.25
TC78402 weakly similar to PIR|B84853|B84853 hypothetical protein... 32 0.25
TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~un... 32 0.43
BF641220 weakly similar to PIR|G86203|G86 probable N-arginine di... 30 0.95
>TC92485 similar to GP|21592663|gb|AAM64612.1 unknown {Arabidopsis
thaliana}, partial (63%)
Length = 704
Score = 430 bits (1105), Expect = e-121
Identities = 216/234 (92%), Positives = 226/234 (96%)
Frame = +2
Query: 6 EELQFLNIPNILKESISIPKISPKTFYLITLILIFPLSFAILAHSLFTHPLISHIESPFT 65
EELQFLNIPNILKESISIPKISPKTFYLITL LIFPLSFAILAHSLFTHPLIS ++SPFT
Sbjct: 2 EELQFLNIPNILKESISIPKISPKTFYLITLTLIFPLSFAILAHSLFTHPLISQLQSPFT 181
Query: 66 DPAQTSHDWTLLLIIQFFYLIFLFAFSLLSTAAVVFTVASLYTSKPVSFSNTISAIPNVF 125
DP+QTSHDWTLLLIIQFFYLIFLFAFSLLSTAAVVFTVASLYTSKPVSFSNTISAIP VF
Sbjct: 182 DPSQTSHDWTLLLIIQFFYLIFLFAFSLLSTAAVVFTVASLYTSKPVSFSNTISAIPKVF 361
Query: 126 KRLFITFLWVTLLMFCYNFVFILCLVLMVIAVDTDNSVLLFFSVVLIFVLFLVVHVYITA 185
KRLFITFLWVTLLM CYNF+F+LCLVLMV+A+DTDNSVLLFFSVV IF LFLVVHVYITA
Sbjct: 362 KRLFITFLWVTLLMICYNFLFVLCLVLMVVAIDTDNSVLLFFSVVFIFDLFLVVHVYITA 541
Query: 186 LWHLASVVSVLEPLYGFAAMKKSYELLKGRVRYASLLVCGYLFLCAVISGMFSV 239
LWHLASVVSVLEPLYGFAAMKKSYELLKGRVRYA+LLVCGYL +CA ISGMF+V
Sbjct: 542 LWHLASVVSVLEPLYGFAAMKKSYELLKGRVRYAALLVCGYLLICAGISGMFTV 703
>TC79406 weakly similar to PIR|T04879|T04879 hypothetical protein F18F4.50 -
Arabidopsis thaliana, partial (26%)
Length = 1053
Score = 196 bits (499), Expect = 8e-51
Identities = 125/314 (39%), Positives = 168/314 (52%), Gaps = 16/314 (5%)
Frame = +2
Query: 1 MDLAPEELQFLNIPNILKESISIPKISPKTFYLITLILIFPLSFAILAH---SLFTHPLI 57
MD E++QFL KES I K F ITL L+ PLSF L H S I
Sbjct: 122 MDKEQEDMQFLGFFGCYKESFKIIFSWRKIFTQITLTLLLPLSFIFLFHIEVSNLIFKKI 301
Query: 58 SHIESPFTDPAQ-----------TSHDWTLLLIIQFFYLIFLFAFSLLSTAAVVFTVASL 106
H E Q S + LL+ + Y FL FSLLST+A+V+T AS+
Sbjct: 302 QHTEEQIDHTQQGTPKYEKLTDIISSQFITLLLFKLVYFTFLLIFSLLSTSAIVYTTASI 481
Query: 107 YTSKPVSFSNTISAIPNVFKRLFITFLWVTLLMFCYNFV--FILCLVLMVIAVDTDNSVL 164
+TSK VSF + +P V+KRL ITFL F YNF+ F++ L+ + I++ + VL
Sbjct: 482 FTSKDVSFKKVMKIVPKVWKRLMITFLCAYAAFFAYNFITFFVIILLAITISIKSGGIVL 661
Query: 165 LFFSVVLIFVLFLVVHVYITALWHLASVVSVLEPLYGFAAMKKSYELLKGRVRYASLLVC 224
L F ++ F+ F VY+T +W LASVV+VLE YG AM KS EL+KG++ + +
Sbjct: 662 LIFIMIFYFIGF----VYLTLIWQLASVVTVLEDFYGIKAMVKSKELIKGKMGLSIFIFL 829
Query: 225 GYLFLCAVISGMFSVIVVHGGDGYDVFSRIFIGGFLVGVLVIVNLVGLLVQSVFYYVCKS 284
+I +F V+VV+G + R G L + LVGL++Q+V Y VCKS
Sbjct: 830 VLNVSFVLIRLVFKVVVVNGRWCFGYVDRTGYGILCFLFLSCLFLVGLVLQTVLYLVCKS 1009
Query: 285 YHHQEIDKSALHDH 298
YHH+ IDKSAL DH
Sbjct: 1010YHHENIDKSALADH 1051
>BE998132 homologue to GP|21592663|gb| unknown {Arabidopsis thaliana},
partial (16%)
Length = 685
Score = 112 bits (281), Expect = 1e-25
Identities = 53/55 (96%), Positives = 54/55 (97%)
Frame = -2
Query: 266 IVNLVGLLVQSVFYYVCKSYHHQEIDKSALHDHLGGYLGEYVPLKSSIQMENLDI 320
IVNLVGLLVQSVFYYVCKSYHHQ IDKSALHDHLGGYLGEYVPLKSSIQMENLD+
Sbjct: 684 IVNLVGLLVQSVFYYVCKSYHHQGIDKSALHDHLGGYLGEYVPLKSSIQMENLDV 520
>TC85499 similar to GP|22477284|gb|AAH28621.1 Unknown (protein for
MGC:33579) {Homo sapiens}, partial (5%)
Length = 631
Score = 98.6 bits (244), Expect = 3e-21
Identities = 58/164 (35%), Positives = 84/164 (50%), Gaps = 14/164 (8%)
Frame = +2
Query: 9 QFLNIPNILKESISIPKISPKTFYLITLILIFPLSFAILAHSLFTHPLISHIESPFTDPA 68
QFL + I KES I K F ITL LI PLSF L H ++ L I + +
Sbjct: 2 QFLGLFGIYKESYKIIISWRKIFSQITLTLILPLSFIFLIHIQISNILFRKIMNNTNEIL 181
Query: 69 QTSH--------------DWTLLLIIQFFYLIFLFAFSLLSTAAVVFTVASLYTSKPVSF 114
+T +WT L+ + Y L FSLLST+AVV+T+AS+YT++ V+F
Sbjct: 182 ETQQGTPQYQKLSDIITSEWTTFLLFKLVYFTSLLIFSLLSTSAVVYTIASIYTAREVTF 361
Query: 115 SNTISAIPNVFKRLFITFLWVTLLMFCYNFVFILCLVLMVIAVD 158
+P V+KRL +TFL F YN + IL ++ ++++
Sbjct: 362 KRVFGVVPKVWKRLMVTFLCTFFAFFVYNTLAILVFIIWALSIN 493
>TC77789 weakly similar to GP|6671937|gb|AAF23197.1| unknown protein
{Arabidopsis thaliana}, partial (32%)
Length = 1149
Score = 78.2 bits (191), Expect = 4e-15
Identities = 68/290 (23%), Positives = 131/290 (44%), Gaps = 10/290 (3%)
Frame = +3
Query: 12 NIPNILKESISIPKISPKTFYLITLILIFPLSFAILAHSLFTHPLISHIESPFTDPAQTS 71
N+ N+L ES SI P + ++LI + PLSF L L I F T+
Sbjct: 63 NMWNVLSESKSIVNSQPHHYLTLSLIFLLPLSFLSL--------LFQFISKNFQQQLSTN 218
Query: 72 HDWTLLLIIQFFYLIFLFAFSLLSTAAVVFTVASLYTSKPVSFSNTISAIPNVFKRLFIT 131
+ L + F L + AF + T + ++V ++PV + +I F L T
Sbjct: 219 PTTIISLYLLFIILSCIIAFGAVIT--ITYSVFHACFNRPVKLKEALKSIITSFFPLLAT 392
Query: 132 FLWVTLLMFCYNFVF--ILCLVLMVIA------VDTDNSVLLFFSVVLIFVLFLVVHVYI 183
++ F F+F ++ LVL +I + T+ +++ +VL ++ L + Y+
Sbjct: 393 ATVAFVVFFFIFFLFGLLIGLVLFLITYLGHVDLKTNPNLVALCFMVLFMIVLLPLVTYL 572
Query: 184 TALWHLASVVSVLEPLYGFAAMKKSYELLKG--RVRYASLLVCGYLFLCAVISGMFSVIV 241
++ V+E +GF +++S++L+KG R+ +++L+ G+L + + +S+++
Sbjct: 573 LVNLSFMKIIVVVESSWGFEPLRRSWKLVKGMKRLVLSTILLFGFLEVILLKISCYSLVL 752
Query: 242 VHGGDGYDVFSRIFIGGFLVGVLVIVNLVGLLVQSVFYYVCKSYHHQEID 291
V + +L I+ L ++V +V Y CK H + D
Sbjct: 753 VL---------------VISPILAILMLYNIVVNTVLYIHCKEKHGEVAD 857
>TC81307 weakly similar to GP|4335772|gb|AAD17449.1| unknown protein
{Arabidopsis thaliana}, partial (31%)
Length = 1171
Score = 76.6 bits (187), Expect = 1e-14
Identities = 73/304 (24%), Positives = 130/304 (42%), Gaps = 28/304 (9%)
Frame = +1
Query: 11 LNIPNILKESISIPKISPKTFYLITLILIFPLSFAILAHSLFTHPLISHIESP------- 63
L + +IL ES I + F +++I + PLSF+++ H L + SP
Sbjct: 88 LKLWSILSESKRIINAHSRHFLALSVIFLLPLSFSLIVSPTIFHLLSTPSNSPTIHILLR 267
Query: 64 -----------------FTDPAQTSHDWTLLLIIQFFYLIFLFAFSLLSTAAVVFTVASL 106
FT TS D L L + L FLF SL S A + +V
Sbjct: 268 QATSLLSLQLRTEEDIRFTPKTTTSLDLPLPLPLLLLPLFFLFFLSLCSLATITHSVFHG 447
Query: 107 YTSKPVSFSNTISAIPNVFKRLFITFLWVTLLMFCYNFVFILCLVLMVIAVDTDNSVLLF 166
+ +PV +T++++ + F L T T+L F L + L+ ++ + + ++
Sbjct: 448 FFGRPVKLLSTVTSLLSSFLPLLTT----TILSHLILFTISLPIPLLRLSF-SFSEIIRT 612
Query: 167 FSVVLIFVLFLVVHVYITALWHLASVVSVLEPLYGFAAMKKSYELLKGRVRYASLLVCGY 226
S +L+ L L++ YI W L+SV++++E YG +++S L+ G ++Y C +
Sbjct: 613 VSAILVLALSLLI-FYIRISWTLSSVIAIVENTYGIQPLRRSAHLMNG-MKYTG-ASCFF 783
Query: 227 LFL----CAVISGMFSVIVVHGGDGYDVFSRIFIGGFLVGVLVIVNLVGLLVQSVFYYVC 282
F + SG VV + ++ + V+ ++ L +V Y C
Sbjct: 784 FFASLEGIMLWSGFLLARVVSDSGSWRDWAFMVQIVLTSAVMTVLMLYNAAANTVLYMYC 963
Query: 283 KSYH 286
K+ H
Sbjct: 964 KALH 975
>TC81606 similar to EGAD|129691|138422 fibroblast growth factor 10 {Gallus
gallus}, partial (8%)
Length = 840
Score = 67.0 bits (162), Expect = 9e-12
Identities = 60/258 (23%), Positives = 120/258 (46%)
Frame = +3
Query: 12 NIPNILKESISIPKISPKTFYLITLILIFPLSFAILAHSLFTHPLISHIESPFTDPAQTS 71
+I N+L ESI I P+ + ++LI + P SF IL L +I H++
Sbjct: 15 SIWNVLSESIHIINSQPRHYLTLSLIFLLPFSFLILISKL----IIKHLQ---------Q 155
Query: 72 HDWTLLLIIQFFYLIFLFAFSLLSTAAVVFTVASLYTSKPVSFSNTISAIPNVFKRLFIT 131
+++ + +LI FS + A+ ++V + + ++PV I +I F L T
Sbjct: 156 QQPPIIISLYLLFLILSSIFSYSAFIAITYSVYNAFFNQPVKLKEAIKSITTSFFPLLAT 335
Query: 132 FLWVTLLMFCYNFVFILCLVLMVIAVDTDNSVLLFFSVVLIFVLFLVVHVYITALWHLAS 191
++ + F F+F+ + +L + ++L+F+ +L V++ L
Sbjct: 336 EAIISPIFFVNFFLFVYIKSYSYL------FMLFYMVLMLVFMSYLEVNL------GLVK 479
Query: 192 VVSVLEPLYGFAAMKKSYELLKGRVRYASLLVCGYLFLCAVISGMFSVIVVHGGDGYDVF 251
V+ V+E +G +K+S++L+KG + +L YLF F +++ GY++
Sbjct: 480 VIVVVESSWGLEPLKRSWKLVKGMRKL--VLSIFYLF-------GFLEVILRWISGYNLV 632
Query: 252 SRIFIGGFLVGVLVIVNL 269
IF+ + +L++ N+
Sbjct: 633 -LIFVISPIQAMLMLYNI 683
>TC78636
Length = 996
Score = 45.4 bits (106), Expect = 3e-05
Identities = 66/275 (24%), Positives = 114/275 (41%), Gaps = 29/275 (10%)
Frame = +2
Query: 11 LNIPNILKESISIPKISPKTFYLITLILIFPLSFAILA-HSLFTHPLISH----IESPFT 65
+++ +IL E+++I F + TL+ P F ++ +LF L+ + PF
Sbjct: 50 IDVLDILGETLTI-YFKNINFIIFTLLTSLPFFFLMVYFETLFQQTLVQTPKIILPLPFH 226
Query: 66 DPAQT--------------SHDWTLLLI-IQFFYLIFLFAFSLLSTAAVVFTVASLYTSK 110
+ Q S D+ LLI + F Y I L S + + L +
Sbjct: 227 ERNQLRFYSVIDIYNGPSFSKDYLPLLIQLGFIYTIPLHFLEFCSKVLTMDLASKLIINS 406
Query: 111 PVS--------FSNTISAIPNVFKRLFITFLWVTLLMFCYNFVFILCLVLMVIAVDTDNS 162
+ F N+I + K FIT L+ L C F + I T
Sbjct: 407 EENPKMSLKHMFQNSIDV--STMKGTFITSLYTLALSNCLLIAFPWTVSNCFIF--TRWC 574
Query: 163 VLLFFSVVLIFV-LFLVVHVYITALWHLASVVSVLEPLYGFAAMKKSYELLKGRVRYASL 221
++F S+ I V L++++ +A+W+++ VVSVLE +YG A+K SY G + +
Sbjct: 575 YIIFGSICFIAVGKLLMIYLECSAIWNMSIVVSVLEGIYGTGALKGSYYFSSGNHKRGLV 754
Query: 222 LVCGYLFLCAVISGMFSVIVVHGGDGYDVFSRIFI 256
++ + V+ M + G G +F +I I
Sbjct: 755 MMLVFFVFGGVLRLMCIYFECYKG-GSGIFIQIGI 856
>BM814285 weakly similar to GP|9295718|gb| T24P13.3 {Arabidopsis thaliana},
partial (36%)
Length = 376
Score = 45.4 bits (106), Expect = 3e-05
Identities = 30/125 (24%), Positives = 60/125 (48%), Gaps = 6/125 (4%)
Frame = +3
Query: 107 YTSKPVSFSNTISAIPNVFKRLFITFLWVT------LLMFCYNFVFILCLVLMVIAVDTD 160
Y+ K S + ++++ T++W + MFC F+ C L V+ +
Sbjct: 6 YSRKKFDGSKFCVIVAKFWRKILATYMWACTVIVGCITMFCV-FLVAFCSALAVLGFSPN 182
Query: 161 NSVLLFFSVVLIFVLFLVVHVYITALWHLASVVSVLEPLYGFAAMKKSYELLKGRVRYAS 220
++ ++ +++ ++F VV + +A V+SVLE + G AM +S L+KG+ +
Sbjct: 183 ---VVVYAALMVGLVFSVVFANAIIICSVALVISVLEDVSGAQAMLRSSILIKGQTQVGL 353
Query: 221 LLVCG 225
L+ G
Sbjct: 354 LIYLG 368
>TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana tabacum},
partial (13%)
Length = 663
Score = 43.1 bits (100), Expect = 1e-04
Identities = 35/116 (30%), Positives = 55/116 (47%), Gaps = 7/116 (6%)
Frame = -2
Query: 76 LLLIIQFFYLIFLFAFSLLSTAAVVFTVASLYTSKPVSFSNTISAIPNVFKRLFITFLWV 135
LLL+ F LI LF S+L+ V V L +F+ FL
Sbjct: 509 LLLLFLFLILILLFLTSILTFIIVFILVLFLLL-------------------VFLRFLLF 387
Query: 136 TLLMFCYN-----FVFILCLVLMVIAVD--TDNSVLLFFSVVLIFVLFLVVHVYIT 184
T+L+F + F+F+L L+L+ I + T N + L F +L+FVL ++ +YI+
Sbjct: 386 TILVFVFFSLLLLFLFLLLLILLFITIFFFTLNILFLLFFFLLLFVLLILTLIYIS 219
Score = 34.7 bits (78), Expect = 0.050
Identities = 31/125 (24%), Positives = 57/125 (44%), Gaps = 4/125 (3%)
Frame = -2
Query: 76 LLLIIQFFYLIFLFAFSLLSTAAVVFTVASLYTSKPVSFSNTISAIPNVFKRLFITFLWV 135
LLL + FF+L+ LF F Y + P + I + I L
Sbjct: 617 LLLFLVFFFLLLLFTFFR----------GFFYINIPYRYFTLFFLILLLLFLFLILILLF 468
Query: 136 TLLMFCYNFVFILCLVLMVIAVD---TDNSVLLFFSVVLIFV-LFLVVHVYITALWHLAS 191
+ + VFIL L L+++ + V +FFS++L+F+ L L++ ++IT + +
Sbjct: 467 LTSILTFIIVFILVLFLLLVFLRFLLFTILVFVFFSLLLLFLFLLLLILLFITIFFFTLN 288
Query: 192 VVSVL 196
++ +L
Sbjct: 287 ILFLL 273
>TC83625 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana tabacum},
partial (9%)
Length = 908
Score = 41.6 bits (96), Expect = 4e-04
Identities = 31/89 (34%), Positives = 50/89 (55%), Gaps = 9/89 (10%)
Frame = -2
Query: 105 SLYTSKPVSFSNTISAIPNVFKRL--FITFLWVTLLMFCYN-----FVFILCLVLMVIAV 157
SL + P SFS++I V L F+ FL T+L+F + F+F+L L+L+ I +
Sbjct: 877 SLTDTLPSSFSSSIIVFILVLFLLLVFLRFLLFTILVFVFFSLLLLFLFLLLLILLFITI 698
Query: 158 D--TDNSVLLFFSVVLIFVLFLVVHVYIT 184
T N + L F +L+FVL ++ +YI+
Sbjct: 697 FFFTLNILFLLFFFLLLFVLLILTLIYIS 611
Score = 38.5 bits (88), Expect = 0.003
Identities = 35/126 (27%), Positives = 62/126 (48%)
Frame = -2
Query: 57 ISHIESPFTDPAQTSHDWTLLLIIQFFYLIFLFAFSLLSTAAVVFTVASLYTSKPVSFSN 116
+S I TD +S ++++ I +L+ +F LL T +VF SL + F
Sbjct: 895 VSSISISLTDTLPSSFSSSIIVFILVLFLLLVFLRFLLFTI-LVFVFFSLL----LLFLF 731
Query: 117 TISAIPNVFKRLFITFLWVTLLMFCYNFVFILCLVLMVIAVDTDNSVLLFFSVVLIFVLF 176
+ I LFIT + TL + F F+L VL+++ + + VLL ++++ LF
Sbjct: 730 LLLLI-----LLFITIFFFTLNILFLLFFFLLLFVLLILTLIYISFVLL---LLILLFLF 575
Query: 177 LVVHVY 182
L++ +Y
Sbjct: 574 LIIFLY 557
>TC80288 weakly similar to PIR|T06029|T06029 hypothetical protein T28I19.100
- Arabidopsis thaliana, partial (14%)
Length = 1460
Score = 39.7 bits (91), Expect = 0.002
Identities = 55/198 (27%), Positives = 87/198 (43%), Gaps = 3/198 (1%)
Frame = -1
Query: 76 LLLIIQFFYLIFLFAFSLLSTAAVVFTVASLYTSKPVSFSNTISAIPNVFKRLFITFLWV 135
L LII F+ +FLF +L+ T + + + N S I ++F TF ++
Sbjct: 743 LRLIITLFFFVFLFIHNLILTIIICL*FLMII----FIYLNISSFIFHLF-----TFAFI 591
Query: 136 TLLMFC--YNFVFILCLVLMVIAVDTDNSVLLFFSVVLI-FVLFLVVHVYITALWHLASV 192
LL FC F FIL + V T FFS VL+ F FL++ + T + H+
Sbjct: 590 FLLHFCSMLIFFFILFFSPFMHIVFT-----FFFSFVLVSFFSFLLLFIVFTFILHIVYN 426
Query: 193 VSVLEPLYGFAAMKKSYELLKGRVRYASLLVCGYLFLCAVISGMFSVIVVHGGDGYDVFS 252
+ + + F M +L+C L V+S F ++ G + F
Sbjct: 425 MFI----FLFFLM-------------PFILICFNLTRMKVLSTKFQNLISLGSNRQ--FR 303
Query: 253 RIFIGGFLVGVLVIVNLV 270
IFI FL+ V++++NLV
Sbjct: 302 IIFI-KFLLFVVIVLNLV 252
>TC80666 similar to PIR|E85112|E85112 probable methyltransferase [imported]
- Arabidopsis thaliana, partial (39%)
Length = 1616
Score = 35.0 bits (79), Expect = 0.039
Identities = 34/121 (28%), Positives = 53/121 (43%), Gaps = 12/121 (9%)
Frame = +1
Query: 70 TSHDWTLLL------IIQFFYLIFLFAFSLLSTAAVVFTVASLYTSKPVSFSN-TISAIP 122
TSH WT L +I F L FL FSLLST ++ P+S IS +P
Sbjct: 262 TSHTWTCFLLYNPLYLILFLLLTFL*KFSLLSTTVLLLL-------HPLSLLL*PISTLP 420
Query: 123 NVFKRLFITFLWVTLLMFCYNFVFILCLVL-----MVIAVDTDNSVLLFFSVVLIFVLFL 177
N ++ L +T+++ +L L + +V+ D +L F V +F L +
Sbjct: 421 NHLSNNYLKTLLITIIIIFKTLKSLLLLKIQNLRKLVLCPLFDQWLLFVF*VEYLFHLLI 600
Query: 178 V 178
+
Sbjct: 601 L 603
>TC86635 weakly similar to GP|9758600|dbj|BAB09233.1
gene_id:MDF20.10~pir||T08929~similar to unknown protein
{Arabidopsis thaliana}, partial (32%)
Length = 1963
Score = 33.1 bits (74), Expect = 0.15
Identities = 40/125 (32%), Positives = 62/125 (49%), Gaps = 9/125 (7%)
Frame = -1
Query: 63 PFTDPAQTSHDWTLLLIIQFFYLIFLFAFSLLSTAAVVFTVASLYTSKPVSFSNTISAIP 122
PF+ TS W+L L Q +L+F F F L++ F +L+ S T++ +
Sbjct: 334 PFSHWTITS--WSLWLW-QLQFLLFDFHFLLIN-----FPSGTLF*S-----FLTLTTLG 194
Query: 123 NVFKRLFITFLWVTLLMF---CYNFVF------ILCLVLMVIAVDTDNSVLLFFSVVLIF 173
+F FI + V LL+ C++F+ +LCL + V T +LL+FS +LI
Sbjct: 193 LIFFF*FINSILVGLLLLLRLCFSFLLFIALFLLLCLFFRFLIVITF-FLLLYFSFLLII 17
Query: 174 VLFLV 178
LFLV
Sbjct: 16 TLFLV 2
>TC79220 similar to GP|2149570|gb|AAB58577.1| MAP kinase kinase protein
DdMEK1 {Dictyostelium discoideum}, partial (6%)
Length = 1162
Score = 33.1 bits (74), Expect = 0.15
Identities = 21/73 (28%), Positives = 40/73 (54%), Gaps = 5/73 (6%)
Frame = -1
Query: 130 ITFLWVTLLMFCYNFVFILCLVLMVIAVDTDNSVLLFFSVVLIFVLFLVVHVYITA---- 185
I FLW + + F+F L LVL+ I V VL+ ++V+I V+ +++ V+ +
Sbjct: 832 IKFLWFNIKLML--FIFQLLLVLVTIVVAMVILVLIVVTIVIIIVVPVIIVVHSSR*ITI 659
Query: 186 -LWHLASVVSVLE 197
W + +V+++E
Sbjct: 658 HCWIVLFIVNIIE 620
>TC84353 similar to GP|20466332|gb|AAM20483.1 putative protein {Arabidopsis
thaliana}, partial (9%)
Length = 602
Score = 32.7 bits (73), Expect = 0.19
Identities = 25/82 (30%), Positives = 41/82 (49%), Gaps = 5/82 (6%)
Frame = -3
Query: 31 FYLITLILIFPLSFAILAHSLFTHPLISHIESPFTDPAQTSHDWTLLLIIQF-----FYL 85
F+L +FPL+ I+ + H LI I + T P Q + T + + F FYL
Sbjct: 303 FFLFINFNVFPLTLIIITLTPLQHSLILRIITIHT-PHQLNFRLTPPISLHFQTHLCFYL 127
Query: 86 IFLFAFSLLSTAAVVFTVASLY 107
+FLF L+ ++FT+ +L+
Sbjct: 126 LFLFQLHLV----IIFTLYTLF 73
>BG586161 similar to GP|21554129|gb| putative thaumatin {Arabidopsis
thaliana}, partial (6%)
Length = 754
Score = 32.3 bits (72), Expect = 0.25
Identities = 32/107 (29%), Positives = 43/107 (39%), Gaps = 12/107 (11%)
Frame = -3
Query: 63 PFTDPAQTSHDWTLLLIIQFFYLIFLFAFSL---LSTAAV---------VFTVASLYTSK 110
P P H T +II+ I + SL L T+A F+ SLY SK
Sbjct: 326 PSASPGTAKHAATDAVIIRVETNISVSTMSLNVGLGTSADKYCGTMLLGTFSALSLYLSK 147
Query: 111 PVSFSNTISAIPNVFKRLFITFLWVTLLMFCYNFVFILCLVLMVIAV 157
P SFS + S L +C +F+FI C VL+ A+
Sbjct: 146 PFSFSFSFS-----------------FLCWCNSFLFIFCSVLVEFAL 57
>TC78402 weakly similar to PIR|B84853|B84853 hypothetical protein At2g42370
[imported] - Arabidopsis thaliana, partial (14%)
Length = 1982
Score = 32.3 bits (72), Expect = 0.25
Identities = 14/39 (35%), Positives = 28/39 (70%)
Frame = -2
Query: 161 NSVLLFFSVVLIFVLFLVVHVYITALWHLASVVSVLEPL 199
N +L+FF++VL+F+L L++ + L+HL +V ++ P+
Sbjct: 889 NIMLIFFNIVLLFLLLLMLFFFSLILFHLFTVDTLPIPV 773
>TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~unknown
protein {Arabidopsis thaliana}, partial (46%)
Length = 2077
Score = 31.6 bits (70), Expect = 0.43
Identities = 23/106 (21%), Positives = 49/106 (45%), Gaps = 2/106 (1%)
Frame = -1
Query: 75 TLLLIIQFFYLIFLFAFSLLSTAAVVFTVASLYTSKPVSFSNTISAIPNVFKRLFITFLW 134
T LL F+ +F+ LL+ ++F + +++ F I N F+ F+
Sbjct: 703 TSLLEFIIFFTVFISFIPLLAFLVIIFLLL*IFSQ----FIYFQKFILNSTTFNFLFFIL 536
Query: 135 VTLLMFCYNFVFILCLVLMVIAVDTDNSVLLF--FSVVLIFVLFLV 178
+ + + F+F++ +V + +LLF F +++I + FL+
Sbjct: 535 LIVFILTSFFIFVIFFFEIVFTFTSSFCLLLFNTFKIIIIIIQFLI 398
>BF641220 weakly similar to PIR|G86203|G86 probable N-arginine dibasic
convertase [imported] - Arabidopsis thaliana, partial
(5%)
Length = 634
Score = 30.4 bits (67), Expect = 0.95
Identities = 14/45 (31%), Positives = 28/45 (62%), Gaps = 1/45 (2%)
Frame = -1
Query: 144 FVFILCLVLMVIAVDTDNSVLLFFSVVLIFVL-FLVVHVYITALW 187
F+FIL +++I +++FF ++LIF++ FL++ + T W
Sbjct: 463 FIFILXFFILIIFT----LIIIFFIILLIFIIFFLIIFIN*TIFW 341
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.332 0.146 0.441
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,728,272
Number of Sequences: 36976
Number of extensions: 204508
Number of successful extensions: 2716
Number of sequences better than 10.0: 103
Number of HSP's better than 10.0 without gapping: 2643
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2699
length of query: 320
length of database: 9,014,727
effective HSP length: 96
effective length of query: 224
effective length of database: 5,465,031
effective search space: 1224166944
effective search space used: 1224166944
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (22.0 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146651.4


