

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146651.2 - phase: 0
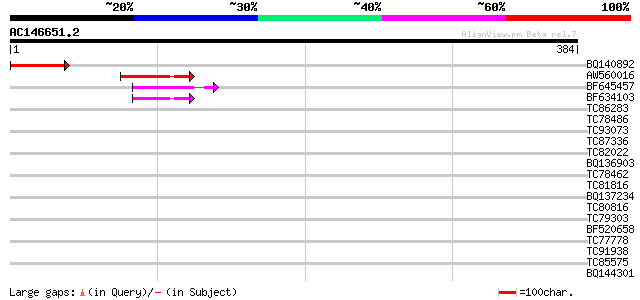
(384 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BQ140892 84 9e-17
AW560016 similar to GP|8778713|gb T25N20.3 {Arabidopsis thaliana... 62 5e-10
BF645457 similar to PIR|T52301|T5 GYMNOS/PICKLE protein [importe... 48 6e-06
BF634103 weakly similar to GP|20260434|gb putative PHD-type zinc... 43 2e-04
TC86283 EGAD|127388|135851 Alfin-1 {Medicago sativa}, partial (97%) 37 0.017
TC78486 similar to GP|21592780|gb|AAM64729.1 nucleic acid bindin... 36 0.022
TC93073 weakly similar to GP|14164467|dbj|BAB55718. hypothetical... 35 0.037
TC87336 similar to GP|10178138|dbj|BAB11550. nucleic acid bindin... 35 0.037
TC82022 similar to GP|6648214|gb|AAF21212.1| unknown protein {Ar... 35 0.049
BQ136903 weakly similar to GP|3878854|emb| cDNA EST yk148b2.5 co... 35 0.063
TC78462 similar to GP|21592780|gb|AAM64729.1 nucleic acid bindin... 32 0.31
TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana taba... 32 0.41
BQ137234 similar to EGAD|35719|3709 hypothetical protein {Burkho... 31 0.92
TC80816 homologue to GP|10178138|dbj|BAB11550. nucleic acid bind... 30 1.6
TC79303 weakly similar to GP|8919075|emb|CAA66109.2 specific tis... 30 1.6
BF520658 PIR|H96667|H96 AP2-containing DNA-binding protein 5168... 30 1.6
TC77778 similar to PIR|T07780|T07780 remorin - potato, partial (... 30 2.0
TC91938 similar to PIR|E96612|E96612 probable transcription fact... 30 2.0
TC85575 similar to PIR|D86410|D86410 protein F3M18.16 [imported]... 29 2.7
BQ144301 similar to GP|15277263|db alternative name: G2~unknown ... 29 3.5
>BQ140892
Length = 273
Score = 84.0 bits (206), Expect = 9e-17
Identities = 40/40 (100%), Positives = 40/40 (100%)
Frame = +1
Query: 1 MKKESKSSAPTMLNKNWVLKRKRRKLPIGPDQSSGKEQSN 40
MKKESKSSAPTMLNKNWVLKRKRRKLPIGPDQSSGKEQSN
Sbjct: 154 MKKESKSSAPTMLNKNWVLKRKRRKLPIGPDQSSGKEQSN 273
>AW560016 similar to GP|8778713|gb T25N20.3 {Arabidopsis thaliana}, partial
(13%)
Length = 657
Score = 61.6 bits (148), Expect = 5e-10
Identities = 24/50 (48%), Positives = 34/50 (68%)
Frame = +1
Query: 76 GHDGYFYECVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
G+D C IC GG+L+CCD CP T+H CLD ++ +P G+W+CP+C
Sbjct: 313 GNDPNDDTCGICGDGGDLICCDGCPSTFHQSCLD--IQMLPPGEWRCPNC 456
>BF645457 similar to PIR|T52301|T5 GYMNOS/PICKLE protein [imported] -
Arabidopsis thaliana, partial (9%)
Length = 560
Score = 48.1 bits (113), Expect = 6e-06
Identities = 22/58 (37%), Positives = 29/58 (49%)
Frame = +2
Query: 84 CVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSCFEENDQLKPLNNLDSI 141
C C G+LL C +C YH CL PPLK W+CP C + PL ++D +
Sbjct: 254 CQACGESGDLLSCATCNYAYHSSCLLPPLKGPAPDNWRCPEC------VTPLIDIDKL 409
>BF634103 weakly similar to GP|20260434|gb putative PHD-type zinc finger
protein {Arabidopsis thaliana}, partial (3%)
Length = 369
Score = 42.7 bits (99), Expect = 2e-04
Identities = 17/42 (40%), Positives = 22/42 (51%)
Frame = +3
Query: 84 CVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
C++C GGNL CD CPR +H +C + P CP C
Sbjct: 72 CIVCWDGGNLFLCDGCPRAFHKECAS--VSSTPRRGRYCPIC 191
>TC86283 EGAD|127388|135851 Alfin-1 {Medicago sativa}, partial (97%)
Length = 2105
Score = 36.6 bits (83), Expect = 0.017
Identities = 27/113 (23%), Positives = 49/113 (42%), Gaps = 7/113 (6%)
Frame = +3
Query: 20 KRKRRKLPIGPDQSSGKEQSNGKEDNSVASESSRSASAKRMLKTEEGTAQFSSKKKGHDG 79
K+ + +L + S+ K +S+GK S + SA + + G + ++G
Sbjct: 1536 KQSKDQLTAHNNGSNSKYKSSGKSRQSESQTKGVKMSAPVKEEVDSGEEEEDDDEQGAT- 1712
Query: 80 YFYECVICDLGGNL------LCCDSCPRTYHLQCLD-PPLKRIPMGKWQCPSC 125
C C G N +CCD C + +H +C+ P K + +++CP C
Sbjct: 1713 ----CGAC--GDNYGTDEFWICCDMCEKWFHGKCVKITPAKAEHIKQYKCPGC 1853
>TC78486 similar to GP|21592780|gb|AAM64729.1 nucleic acid binding
protein-like {Arabidopsis thaliana}, partial (89%)
Length = 1089
Score = 36.2 bits (82), Expect = 0.022
Identities = 25/114 (21%), Positives = 49/114 (42%), Gaps = 8/114 (7%)
Frame = +3
Query: 29 GPDQSSGKEQSNGKEDNSVASESSRSASAKRMLKTEEGTAQFSSK-------KKGHDGYF 81
G + KE+ + NS+ S+S A + K + A+ + +G
Sbjct: 486 GSAKKQTKEKPSVSSHNSIKSKSGSKARGSELAKYSKPPAKEDDEGVDDEEEDQGECAAC 665
Query: 82 YECVICDLGGNLLCCDSCPRTYHLQCLD-PPLKRIPMGKWQCPSCFEENDQLKP 134
E + +CCD C + YH +C+ P + + +++CP+C N +++P
Sbjct: 666 GESYVSASDEFWICCDICEKWYHGKCVKITPARAEHIKQYKCPAC--NNKRVRP 821
>TC93073 weakly similar to GP|14164467|dbj|BAB55718. hypothetical protein
{Oryza sativa (japonica cultivar-group)}, partial (18%)
Length = 776
Score = 35.4 bits (80), Expect = 0.037
Identities = 24/92 (26%), Positives = 41/92 (44%), Gaps = 12/92 (13%)
Frame = +3
Query: 50 ESSRSASAKRMLKTEEGTAQFSSKKKGHDGYFYECVIC----DLGGNLLCCDSCPRTYHL 105
E S + +L + + + +K++ H GY YEC +C + G ++ C +H
Sbjct: 312 ERSFENQQRNLLSSFKYKKEVVAKEQEHKGYDYECYVCLSVYEEGEDVRKLPKCKHCFHA 491
Query: 106 QCLDP--------PLKRIPMGKWQCPSCFEEN 129
C+D P+ R P+G C + EEN
Sbjct: 492 LCIDMWLYSHFDCPICRTPVGP-LCHNFQEEN 584
>TC87336 similar to GP|10178138|dbj|BAB11550. nucleic acid binding
protein-like {Arabidopsis thaliana}, partial (95%)
Length = 1223
Score = 35.4 bits (80), Expect = 0.037
Identities = 30/99 (30%), Positives = 43/99 (43%), Gaps = 15/99 (15%)
Frame = +3
Query: 42 KEDNSVASESSRSASAKRMLKTEEGTAQFSSKKKGHDGYFYE--------CVICDLGGNL 93
K+ +V S S S KR + +G + + K GY E C C GGN
Sbjct: 624 KDKPTVDSGSKSRGSTKR---SSDGQVKSNPKLVDDQGYEEEEDEHSETLCGSC--GGNY 788
Query: 94 ------LCCDSCPRTYHLQCLD-PPLKRIPMGKWQCPSC 125
+ CD C R YH +C+ P K + +++CPSC
Sbjct: 789 NADEFWIGCDICERWYHGKCVKITPAKAESIKQYKCPSC 905
>TC82022 similar to GP|6648214|gb|AAF21212.1| unknown protein {Arabidopsis
thaliana}, partial (24%)
Length = 1153
Score = 35.0 bits (79), Expect = 0.049
Identities = 15/43 (34%), Positives = 19/43 (43%), Gaps = 3/43 (6%)
Frame = +3
Query: 84 CVICDLGGN---LLCCDSCPRTYHLQCLDPPLKRIPMGKWQCP 123
C C G+ + C C YH CL PP K + G + CP
Sbjct: 756 CEACRRTGDPSKFMFCKRCDGAYHCYCLQPPHKNVSTGPYLCP 884
>BQ136903 weakly similar to GP|3878854|emb| cDNA EST yk148b2.5 comes from
this gene {Caenorhabditis elegans}, partial (9%)
Length = 687
Score = 34.7 bits (78), Expect = 0.063
Identities = 17/45 (37%), Positives = 23/45 (50%), Gaps = 3/45 (6%)
Frame = +2
Query: 84 CVICD-LGGNLLCCDS--CPRTYHLQCLDPPLKRIPMGKWQCPSC 125
C+ C+ LGG LL C CP + H+ C+ K G + CP C
Sbjct: 263 CITCNKLGGELLVCSQTDCPVSVHVTCIGSEPKFDDSGNFFCPYC 397
>TC78462 similar to GP|21592780|gb|AAM64729.1 nucleic acid binding
protein-like {Arabidopsis thaliana}, partial (85%)
Length = 1281
Score = 32.3 bits (72), Expect = 0.31
Identities = 13/42 (30%), Positives = 25/42 (58%), Gaps = 1/42 (2%)
Frame = +3
Query: 94 LCCDSCPRTYHLQCLD-PPLKRIPMGKWQCPSCFEENDQLKP 134
+CCD C + +H +C+ P + + +++CPSC N + +P
Sbjct: 849 ICCDICEKWFHGKCVKITPARAEHIKQYKCPSC-SNNKRARP 971
>TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana tabacum},
partial (13%)
Length = 663
Score = 32.0 bits (71), Expect = 0.41
Identities = 42/178 (23%), Positives = 66/178 (36%), Gaps = 8/178 (4%)
Frame = +3
Query: 199 VLSPVDATCSDKPMDPSLESCMEGTSCADADEKNLNLSPTVAPMDRMSVSPDKEVLSPSK 258
+ + D T S K + ++ ++ S D E + + ++ SV D E
Sbjct: 30 LFTTTDITVSSKMEESNIPIKIDDKSAGDVKEDKVEIEKD---LEIKSVEKDDEKKEKKD 200
Query: 259 ITNLDANDDLLEEKPDLSCDKIPFRKTLVLAITVGGEEMGKRKHKVIGDNANQKKRRTEK 318
D D ++E D DK +K G EE G K D +KK + EK
Sbjct: 201 KEKKDKTD--VDEGKDKK-DKEKKKKEKKEENVKGEEEDGDEKK----DKEKKKKEKKEK 359
Query: 319 GKKVVITPIKSKSGNNKVQTKQKSKTHKISISASKGDVG--------KKKSDAQQKDK 368
GK+ K K G K K K K + +G+ G KK+ ++ +K
Sbjct: 360 GKED-----KDKDGEEKKSKKDKEKKKDKNEDDDEGEDGSKKKKNKDKKEKKKEEDEK 518
>BQ137234 similar to EGAD|35719|3709 hypothetical protein {Burkholderia
cepacia}, partial (6%)
Length = 1184
Score = 30.8 bits (68), Expect = 0.92
Identities = 22/74 (29%), Positives = 34/74 (45%)
Frame = +1
Query: 293 GGEEMGKRKHKVIGDNANQKKRRTEKGKKVVITPIKSKSGNNKVQTKQKSKTHKISISAS 352
GGEE K+K + ++RR E+ +K + + G + KQK +THK
Sbjct: 331 GGEETRKKKKQ-------PQRRRGEREEKGEGEEARKREGPGRTNRKQKRRTHK------ 471
Query: 353 KGDVGKKKSDAQQK 366
+ KKS QQ+
Sbjct: 472 --EESGKKSKQQQR 507
>TC80816 homologue to GP|10178138|dbj|BAB11550. nucleic acid binding
protein-like {Arabidopsis thaliana}, partial (15%)
Length = 358
Score = 30.0 bits (66), Expect = 1.6
Identities = 12/31 (38%), Positives = 18/31 (57%), Gaps = 1/31 (3%)
Frame = +1
Query: 96 CDSCPRTYHLQCLD-PPLKRIPMGKWQCPSC 125
CD C R YH +C+ P K + +++CP C
Sbjct: 25 CDICERWYHGKCVKITPAKAESIKQYKCPQC 117
>TC79303 weakly similar to GP|8919075|emb|CAA66109.2 specific tissue protein
2 {Cicer arietinum}, partial (64%)
Length = 1002
Score = 30.0 bits (66), Expect = 1.6
Identities = 17/41 (41%), Positives = 20/41 (48%)
Frame = +3
Query: 306 GDNANQKKRRTEKGKKVVITPIKSKSGNNKVQTKQKSKTHK 346
GDNA + E K P S GNN+V TK+K K K
Sbjct: 642 GDNAIDNSKTNEVIKDFESRPNISAYGNNEVDTKKKEKAAK 764
>BF520658 PIR|H96667|H96 AP2-containing DNA-binding protein 51686-52693
[imported] - Arabidopsis thaliana, partial (5%)
Length = 538
Score = 30.0 bits (66), Expect = 1.6
Identities = 15/41 (36%), Positives = 22/41 (53%)
Frame = +1
Query: 312 KKRRTEKGKKVVITPIKSKSGNNKVQTKQKSKTHKISISAS 352
+K + EK KK+ +K SG+N TK + KI S+S
Sbjct: 55 QKVKREKAKKIAAKKMKKNSGSNNHDTKSDKNSDKIINSSS 177
>TC77778 similar to PIR|T07780|T07780 remorin - potato, partial (68%)
Length = 1103
Score = 29.6 bits (65), Expect = 2.0
Identities = 36/174 (20%), Positives = 61/174 (34%), Gaps = 21/174 (12%)
Frame = +3
Query: 203 VDATCSDKPMDPSLESCMEGTSCADADEKNLNLSPTVAPMDRMSVSPDKEVLSPSKITNL 262
+++ + P P + T +A +K++ +V P D P V + +
Sbjct: 285 IESESTSNPPPPPASTETTTTPLPEAPKKDVAEEKSVIPQDNNPPPPPPVVDDSKALVIV 464
Query: 263 DANDDLLEEKP--------DLSCDKIPFRKTLVLAITVGGEEMGKRKHK------VIGDN 308
D+ EEKP D ++ K L L E K ++K I
Sbjct: 465 QKTDEAAEEKPKEGGSIDRDAVLTRVATEKRLSLIKAWEESEKSKAENKAQRRLSTITAW 644
Query: 309 ANQKKRRTEKGKKVVITPIKSKSGNNKVQTKQK-------SKTHKISISASKGD 355
N KK E + + ++ K G + K K ++ K I A KG+
Sbjct: 645 ENSKKAAKEAELRKLEEQLEKKKGEYAEKLKNKIAALHKAAEEKKAMIEAKKGE 806
>TC91938 similar to PIR|E96612|E96612 probable transcription factor
F12K22.14 [imported] - Arabidopsis thaliana, partial
(11%)
Length = 685
Score = 29.6 bits (65), Expect = 2.0
Identities = 14/50 (28%), Positives = 21/50 (42%), Gaps = 3/50 (6%)
Frame = +3
Query: 84 CVICDLGGN---LLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSCFEEND 130
C++C + L C +C +H CL M W CP C + +D
Sbjct: 99 CMVCKQKPSETETLHCKTCTTPWHAPCLPVVPTTSEMLDWLCPDCAQPSD 248
>TC85575 similar to PIR|D86410|D86410 protein F3M18.16 [imported] -
Arabidopsis thaliana, partial (15%)
Length = 1225
Score = 29.3 bits (64), Expect = 2.7
Identities = 13/50 (26%), Positives = 23/50 (46%)
Frame = +3
Query: 33 SSGKEQSNGKEDNSVASESSRSASAKRMLKTEEGTAQFSSKKKGHDGYFY 82
+S + Q+N DN ++ S+ TEEG ++ ++ YFY
Sbjct: 555 NSNENQNNNNNDNKYFYNNNNKDSSSNTKFTEEGYNSMENRNNNNNKYFY 704
>BQ144301 similar to GP|15277263|db alternative name: G2~unknown function
{Homo sapiens}, partial (1%)
Length = 1250
Score = 28.9 bits (63), Expect = 3.5
Identities = 15/41 (36%), Positives = 22/41 (53%)
Frame = -3
Query: 310 NQKKRRTEKGKKVVITPIKSKSGNNKVQTKQKSKTHKISIS 350
N +K+ +K K ITP K+ NNK +T +K + IS
Sbjct: 642 NNRKKNQQKNK---ITPTKTPQSNNKPKTSAPNKKRRSPIS 529
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.311 0.129 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,034,446
Number of Sequences: 36976
Number of extensions: 154432
Number of successful extensions: 696
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 688
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 695
length of query: 384
length of database: 9,014,727
effective HSP length: 98
effective length of query: 286
effective length of database: 5,391,079
effective search space: 1541848594
effective search space used: 1541848594
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146651.2


