

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146651.1 - phase: 0
(935 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
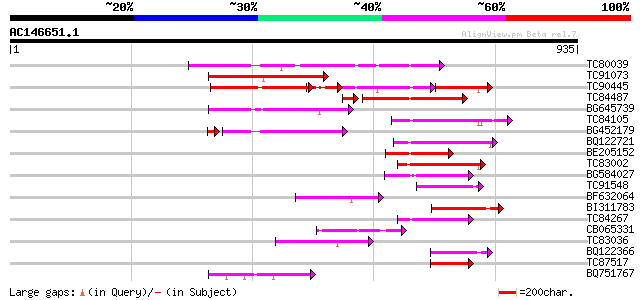
Score E
Sequences producing significant alignments: (bits) Value
TC80039 homologue to GP|6437558|gb|AAF08585.1| putative ATPase (... 250 2e-66
TC91073 similar to PIR|T52301|T52301 GYMNOS/PICKLE protein [impo... 206 4e-53
TC90445 homologue to PIR|C84507|C84507 hypothetical protein At2g... 149 9e-52
TC84487 similar to PIR|T52301|T52301 GYMNOS/PICKLE protein [impo... 186 6e-51
BG645739 similar to PIR|T45808|T4 helicase-like protein - Arabid... 160 3e-39
TC84105 homologue to PIR|C84507|C84507 hypothetical protein At2g... 143 3e-34
BG452179 homologue to GP|15528650|d putative DNA-dependent ATPas... 135 6e-34
BQ122721 similar to GP|8843733|dbj SWI2/SNF2-like protein {Arabi... 131 1e-30
BE205152 similar to GP|19347965|gb putative helicase {Arabidopsi... 111 1e-24
TC83002 similar to GP|14532630|gb|AAK64043.1 putative DNA-bindin... 94 2e-19
BG584027 similar to GP|10178052|db helicase-like transcription f... 79 1e-14
TC91548 similar to PIR|D86185|D86185 hypothetical protein [impor... 78 1e-14
BF632064 similar to PIR|H84432|H84 probable helicase [imported] ... 78 1e-14
BI311783 homologue to PIR|T47587|T4 TATA box binding protein (TB... 74 3e-13
TC84267 weakly similar to PIR|S41478|S41478 DNA repair protein r... 69 6e-12
CB065331 similar to PIR|D83545|D83 probable helicase PA0799 [imp... 68 1e-11
TC83036 similar to PIR|C96637|C96637 hypothetical protein F11P17... 62 1e-09
BQ122366 similar to PIR|T47325|T4 hypothetical protein T12K4.120... 59 9e-09
TC87517 similar to PIR|F84833|F84833 probable SNF2/SWI2 family t... 55 2e-07
BQ751767 similar to SP|O10302|GTA_N Probable global transactivat... 53 6e-07
>TC80039 homologue to GP|6437558|gb|AAF08585.1| putative ATPase (ISW2-like)
{Arabidopsis thaliana}, partial (47%)
Length = 1840
Score = 250 bits (638), Expect = 2e-66
Identities = 166/434 (38%), Positives = 242/434 (55%), Gaps = 12/434 (2%)
Frame = +1
Query: 295 LLEQPKELRGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFK 354
L+ QP ++G + +QL LNWL + + N ILADEMGLGKT+ + + L+
Sbjct: 607 LVTQPSCIQG-KMRDYQLAGLNWLIRLYENGINGILADEMGLGKTLQTISLLGYLHEFRG 783
Query: 355 VSRPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASDPSGLNKKT 414
++ P +V+ P T+GNW+ E + P + V++ G R I++ +
Sbjct: 784 ITGPHMVVAPKSTLGNWMNEIRRFCPVLRAVKFLGSPDERKHIKE------------ELL 927
Query: 415 EAYKFNVLLTSYEMVLADYSHFRGVPWEVLIV--------DEGHRLKNSESKLFSLLNSI 466
A KF+V +TS+EMV+ + F + + E ++N+E L +S+
Sbjct: 928 VAGKFDVCVTSFEMVIKEKPTFPSI*LALCYY**SPSDKK*ELPPVQNNEGI*NKLPDSL 1107
Query: 467 SFQHRVLLTGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERF---NDLTSAEKVDELKKL 523
H PLQNNL E+++LLNFL P F S F+E F + E V +L K+
Sbjct: 1108 *LAH-------PLQNNLHELWSLLNFLLPEIFSSAETFDEWFQISGENDQQEVVQQLHKV 1266
Query: 524 VSPHMLRRLKKDAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSM 583
+ P +LRRLK D + +PPK E ++ V +S +Q +YY+A+L K+ ++ + G ++ +
Sbjct: 1267 LRPFLLRRLKSDVEKGLPPKKETILKVGMSQMQKQYYKALLQKDLEV---VNAGGERKRL 1437
Query: 584 LNIVMQLRKVCNHPYLIPGTEPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLI 643
LNI MQLRK CNHPYL G EP G + I ++ K+ L+ +L L + RVLI
Sbjct: 1438 LNIAMQLRKCCNHPYLFQGAEP--GPPYTTGDHIITSAGKMVLMDKLLPKLKERDSRVLI 1611
Query: 644 FSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFNQDKS-RFVFLLSTRSC 702
FSQMT+LLDILEDYL F Y R+DG+ DR +I FN+ S +FVFLLSTR+
Sbjct: 1612 FSQMTRLLDILEDYL--MFRGYQYCRIDGNTGGDDRDASIEAFNKPGSEKFVFLLSTRAG 1785
Query: 703 GLGINLATADTVII 716
GLGINLATAD VI+
Sbjct: 1786 GLGINLATADVVIL 1827
>TC91073 similar to PIR|T52301|T52301 GYMNOS/PICKLE protein [imported] -
Arabidopsis thaliana, partial (14%)
Length = 627
Score = 206 bits (523), Expect = 4e-53
Identities = 104/207 (50%), Positives = 141/207 (67%), Gaps = 9/207 (4%)
Frame = +3
Query: 329 ILADEMGLGKTISACAFISSLYFEFKVSRPCLVLVPLVTMGNWLAEFALWAPDVNVVQYH 388
ILADEMGLGKTI + AF++SL+ E + P LV+ PL T+ NW EFA WAP +NV+ Y
Sbjct: 3 ILADEMGLGKTIQSIAFLASLFEEGVSAHPHLVVAPLSTLRNWEREFATWAPQMNVIMYV 182
Query: 389 GCAKARAIIRQYEWHASDPSGLNKKTEAY---------KFNVLLTSYEMVLADYSHFRGV 439
G A+AR++IR+YE++ NKK ++ KF+VLLTSYEM+ D + + +
Sbjct: 183 GSAQARSVIREYEFYFPKKLKKNKKKKSLVSESKHDRIKFDVLLTSYEMINLDTTSLKPI 362
Query: 440 PWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLLTGTPLQNNLGEMYNLLNFLQPASFP 499
WE +IVDEGHRLKN +SKLFS L S +HRVLLTGTPLQNNL E++ L++FL F
Sbjct: 363 KWECMIVDEGHRLKNKDSKLFSSLKQYSTRHRVLLTGTPLQNNLDELFMLMHFLDAGKFA 542
Query: 500 SLSAFEERFNDLTSAEKVDELKKLVSP 526
SL F+E F D+ E++ +L K+++P
Sbjct: 543 SLEEFQEEFKDINQEEQISKLHKMLAP 623
>TC90445 homologue to PIR|C84507|C84507 hypothetical protein At2g13370
[imported] - Arabidopsis thaliana, partial (29%)
Length = 1672
Score = 149 bits (377), Expect = 4e-36
Identities = 73/170 (42%), Positives = 110/170 (63%)
Frame = +2
Query: 331 ADEMGLGKTISACAFISSLYFEFKVSRPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGC 390
ADEMGLGKT+ + + + L ++ P LV+VPL T+ NW EF W PD+N++ Y G
Sbjct: 41 ADEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPLSTLSNWAKEFRKWLPDMNIIVYVGT 220
Query: 391 AKARAIIRQYEWHASDPSGLNKKTEAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGH 450
+R + +QYE++ G + KFN LLT+YE++L D + + W L+VDE H
Sbjct: 221 RASREVCQQYEFYNDKKPG-----KPIKFNALLTTYEVILKDKAVLSKIKWNYLMVDEAH 385
Query: 451 RLKNSESKLFSLLNSISFQHRVLLTGTPLQNNLGEMYNLLNFLQPASFPS 500
RLKNSE++L++ L S ++++L+TGTPLQN++ E++ LL+FL P F S
Sbjct: 386 RLKNSEAQLYTSLLEFSTKNKLLITGTPLQNSVEELWALLHFLDPTKFKS 535
Score = 110 bits (274), Expect(3) = 9e-52
Identities = 69/169 (40%), Positives = 99/169 (57%), Gaps = 8/169 (4%)
Frame = +1
Query: 541 PPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIA--QQSMLNIVMQLRKVCNHPY 598
PPK VE+S +Q +YY+ +L +N+Q N+ KG+ Q S+LNIV++L+K CNHP+
Sbjct: 664 PPKLSASFRVEMSPLQKQYYKWILERNFQ---NLNKGVRGNQVSLLNIVVELKKCCNHPF 834
Query: 599 LIPGTE-----PDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIFSQMTKLLDI 653
L + GS E + +S KL +L +L L++ HRVLIFSQM ++LDI
Sbjct: 835 LFESADHGYGGDSGGSDNSKLERIVFSSGKLVILDKLLVRLHETKHRVLIFSQMVRMLDI 1014
Query: 654 LEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFNQDKS-RFVFLLSTRS 701
L YL++ ++R+DGS RQ A+ FN S F FLLSTR+
Sbjct: 1015LAQYLSLR--GFQFQRLDGSTKSELRQQAMEHFNAPGSDDFCFLLSTRA 1155
Score = 99.4 bits (246), Expect(3) = 9e-52
Identities = 58/110 (52%), Positives = 72/110 (64%), Gaps = 17/110 (15%)
Frame = +2
Query: 703 GLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAK 762
GLGINLATADTVII+DSD+NP D+QAM+RAHRIGQ + + +YR V SVEE IL+ AK
Sbjct: 1160 GLGINLATADTVIIFDSDWNPQNDLQAMSRAHRIGQQDVVNIYRFVTSKSVEEDILERAK 1339
Query: 763 KKLMLDQLF----------------KGKSGSQK-EVEDILKWGTEELFND 795
KK++LD L KG S K E+ IL++G EELF +
Sbjct: 1340 KKMVLDHLVIQKLNAEGRLEKKEVKKGGSYFDKNELSAILRFGAEELFKE 1489
Score = 34.7 bits (78), Expect(3) = 9e-52
Identities = 21/59 (35%), Positives = 37/59 (62%)
Frame = +3
Query: 490 LNFLQPASFPSLSAFEERFNDLTSAEKVDELKKLVSPHMLRRLKKDAMQNIPPKTERMV 548
+ F+Q ++ +LS+F E N+L + L + PH+LRR+ KD +++PPK ER++
Sbjct: 540 MKFVQ--NYKNLSSFHE--NELAN------LHMELRPHILRRVIKDVEKSLPPKIERIL 686
>TC84487 similar to PIR|T52301|T52301 GYMNOS/PICKLE protein [imported] -
Arabidopsis thaliana, partial (14%)
Length = 612
Score = 186 bits (471), Expect(2) = 6e-51
Identities = 92/174 (52%), Positives = 129/174 (73%), Gaps = 1/174 (0%)
Frame = +3
Query: 582 SMLNIVMQLRKVCNHPYLIPGTEPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRV 641
S++N+VM+LRK+C H Y++ G EPD + + +++S KL LL M+ L ++GHRV
Sbjct: 96 SLINVVMELRKLCCHAYMLEGVEPDIDDPKEAFKQLLESSGKLHLLDKMMVKLKEQGHRV 275
Query: 642 LIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFN-QDKSRFVFLLSTR 700
LI+SQ +LD+LEDY + + YER+DG V +RQ I RFN ++ SRF FLLSTR
Sbjct: 276 LIYSQFQHMLDLLEDYCS--YKKWHYERIDGKVGGAERQIRIDRFNAKNSSRFCFLLSTR 449
Query: 701 SCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVE 754
+ GLGINLATADTV+IYD D+NPHAD+QAM RAHR+GQ+N++L+Y+L+ R ++E
Sbjct: 450 AGGLGINLATADTVVIYDRDWNPHADLQAMARAHRLGQTNKVLIYKLITRGTIE 611
Score = 34.7 bits (78), Expect(2) = 6e-51
Identities = 16/26 (61%), Positives = 20/26 (76%)
Frame = +2
Query: 550 VELSSIQAEYYRAMLTKNYQILRNIG 575
V+ SS Q EYY+A+LT+NYQIL G
Sbjct: 5 VDSSSKQKEYYKAILTRNYQILTRRG 82
>BG645739 similar to PIR|T45808|T4 helicase-like protein - Arabidopsis
thaliana, partial (17%)
Length = 818
Score = 160 bits (404), Expect = 3e-39
Identities = 87/248 (35%), Positives = 144/248 (57%), Gaps = 10/248 (4%)
Frame = +2
Query: 329 ILADEMGLGKTISACAFISSLYFEFKVSRPCLVLVPLVTMGNWLAEFALWAPDVNVVQYH 388
ILADEMGLGKTI A F++ L E + P L++ P + NW E + P++ V+ Y
Sbjct: 2 ILADEMGLGKTIQAMVFLAHLAEEKNIWGPFLIVAPASVLNNWNEELERFCPELKVLPYW 181
Query: 389 GCAKARAIIRQYEWHASDPSGLNKKTEAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVDE 448
G R ++R+ + +P L ++ KF++L+TSY+++++D +FR V W+ +++DE
Sbjct: 182 GGLSERTVLRK----SMNPKDLYRREA--KFHILITSYQLLVSDEKYFRRVKWQYMVLDE 343
Query: 449 GHRLKNSESKLFSLLNSISFQHRVLLTGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERF 508
+K+S S + L S + ++R+LLTGTP+QNN+ E++ LL+F+ P F S F E F
Sbjct: 344 AQAIKSSNSIRWKTLLSFNCRNRLLLTGTPVQNNMAELWALLHFIMPTLFDSHEQFNEWF 523
Query: 509 N----------DLTSAEKVDELKKLVSPHMLRRLKKDAMQNIPPKTERMVPVELSSIQAE 558
+ + +++ L ++ P MLRR+KKD + + KTE V +LSS Q
Sbjct: 524 SKGIENHAEHGGTLNEHQLNRLHSIIKPFMLRRVKKDVVSELTSKTEITVHCKLSSRQQA 703
Query: 559 YYRAMLTK 566
+Y ++ K
Sbjct: 704 FYSSIKNK 727
>TC84105 homologue to PIR|C84507|C84507 hypothetical protein At2g13370
[imported] - Arabidopsis thaliana, partial (15%)
Length = 834
Score = 143 bits (360), Expect = 3e-34
Identities = 94/219 (42%), Positives = 129/219 (57%), Gaps = 19/219 (8%)
Frame = +1
Query: 630 MLKILYKEGHRVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFNQD 689
+L L++ R+LIFSQM ++LDIL Y+++ ++R+DGS RQ A+ FN
Sbjct: 7 LLVRLHETKFRILIFSQMVRMLDILAQYMSLR--GFQFQRLDGSTKSELRQQAMDHFNAP 180
Query: 690 KSR-FVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLV 748
S F FLLSTR+ GLGINLATADTVII+DSD+NP D+QAM+RAHRIGQ + +YR V
Sbjct: 181 GSDDFCFLLSTRAGGLGINLATADTVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFV 360
Query: 749 VRASVEERILQLAKKKLMLDQLF-------------KGKSG----SQKEVEDILKWGTEE 791
SVEE IL+ AKKK++LD L + K G + E+ IL++G EE
Sbjct: 361 TSKSVEEDILERAKKKMVLDHLVIQKLNAEGKLEKKEAKKGGSFFDKNELSAILRFGAEE 540
Query: 792 LFNDSCALNGKDTSENNNS-NKDEAVAEVEHKHRKRTGG 829
L + N +++ + S + DE + E K GG
Sbjct: 541 LVKEE--RNDEESKKRLLSMDIDEILERAEKVEEKENGG 651
>BG452179 homologue to GP|15528650|d putative DNA-dependent ATPase {Oryza
sativa (japonica cultivar-group)}, partial (19%)
Length = 667
Score = 135 bits (341), Expect(2) = 6e-34
Identities = 77/209 (36%), Positives = 117/209 (55%), Gaps = 4/209 (1%)
Frame = +3
Query: 352 EFK-VSRPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASDPSGL 410
EF+ + P +V+ P T+GNW+ E + P + V++ G + R IR+
Sbjct: 75 EFRGIKGPHMVVAPKSTLGNWMNEIRRFCPILRAVKFLGNPEERRHIRE----------- 221
Query: 411 NKKTEAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQH 470
A KF+V +TS+EM + + S R W +I+DE HR+KN S L + + +
Sbjct: 222 -DLLVAGKFDVCVTSFEMAIKEKSTLRRFSWRYIIIDEAHRIKNENSLLSKTMRIYNTNY 398
Query: 471 RVLLTGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERF---NDLTSAEKVDELKKLVSPH 527
R+L+TGTPLQNNL E+++LLNFL P F S F+E F + E V +L K++ P
Sbjct: 399 RLLITGTPLQNNLHELWSLLNFLLPEIFSSAETFDEWFQISGENDQQEVVQQLHKVLRPF 578
Query: 528 MLRRLKKDAMQNIPPKTERMVPVELSSIQ 556
+LRRLK D + +PPK E ++ V +S +Q
Sbjct: 579 LLRRLKSDVEKGLPPKKETILKVGMSQLQ 665
Score = 27.7 bits (60), Expect(2) = 6e-34
Identities = 12/20 (60%), Positives = 15/20 (75%)
Frame = +2
Query: 327 NVILADEMGLGKTISACAFI 346
N ILADEMGLGKT+ + +
Sbjct: 2 NGILADEMGLGKTLQTISLM 61
>BQ122721 similar to GP|8843733|dbj SWI2/SNF2-like protein {Arabidopsis
thaliana}, partial (19%)
Length = 650
Score = 131 bits (329), Expect = 1e-30
Identities = 83/180 (46%), Positives = 108/180 (59%), Gaps = 9/180 (5%)
Frame = +1
Query: 634 LYKEGHRVLIFSQMTKLLDILEDYLNIEFGPKTYE--RVDGSVSVTDRQTAIARFNQDKS 691
L+ H+VLIFSQ TK+LDI++ Y F K +E R+DGSV + DR+ I FN S
Sbjct: 1 LFARNHKVLIFSQWTKVLDIMDYY----FSEKGFEVCRIDGSVKLDDRKRQIQDFNDTTS 168
Query: 692 RF-VFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVR 750
+FLLSTR+ GLGINL ADT I+YDSD+NP D+QAM+R HRIGQ+ + VYRL
Sbjct: 169 NCRIFLLSTRAGGLGINLTAADTCILYDSDWNPQMDLQAMDRCHRIGQTKPVHVYRLATA 348
Query: 751 ASVEERILQLAKKKLMLDQLF--KGKSGSQKEVEDILKWGTEE----LFNDSCALNGKDT 804
SVE R+L+ A KL L+ + KG+ ++ I+ EE L D L DT
Sbjct: 349 QSVEGRMLKRAFSKLKLEHVVIEKGQFHQERTKPSIMDEMEEEDVLALLRDEEPLRTNDT 528
>BE205152 similar to GP|19347965|gb putative helicase {Arabidopsis thaliana},
partial (27%)
Length = 626
Score = 111 bits (277), Expect = 1e-24
Identities = 60/111 (54%), Positives = 76/111 (68%)
Frame = +1
Query: 621 SAKLTLLHSMLKILYKEGHRVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQ 680
S KL L +LK L HRVL+F+QMTK+L+ILEDY+N + Y R+DGS S+ DR+
Sbjct: 298 SGKLQTLDILLKRLRAGNHRVLLFAQMTKMLNILEDYMN--YRKYKYCRLDGSTSIHDRR 471
Query: 681 TAIARFNQDKSRFVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMN 731
+ F FVFLLSTR+ GLGINL ADTVI Y+SD+NP D+QAM+
Sbjct: 472 DMVRDFQHRSDIFVFLLSTRAGGLGINLTAADTVIFYESDWNPTLDLQAMD 624
>TC83002 similar to GP|14532630|gb|AAK64043.1 putative DNA-binding protein
{Arabidopsis thaliana}, partial (18%)
Length = 1088
Score = 94.4 bits (233), Expect = 2e-19
Identities = 56/150 (37%), Positives = 92/150 (61%), Gaps = 5/150 (3%)
Frame = +3
Query: 640 RVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFNQDKSRFVFLLST 699
+ +IFSQ T +LD++E ++E Y R+DG +++T R A+ FN D V L+S
Sbjct: 453 KAIIFSQWTSMLDLVET--SMEQSGVKYRRLDGRMTLTARDRAVKDFNTDPEITVMLMSL 626
Query: 700 RSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQ 759
++ LG+N+ A VI+ D +NP + QA++RAHRIGQ+ + V R+ ++ +VE+RIL
Sbjct: 627 KAGNLGLNMVAACHVILLDLWWNPTTEDQAIDRAHRIGQTRPVTVTRITIKDTVEDRILA 806
Query: 760 L-AKKKLMLDQLF----KGKSGSQKEVEDI 784
L +K+ M+ F G SG++ V+D+
Sbjct: 807 LQEEKRKMVASAFGEDHAGGSGTRLTVDDL 896
>BG584027 similar to GP|10178052|db helicase-like transcription factor-like
protein {Arabidopsis thaliana}, partial (15%)
Length = 830
Score = 78.6 bits (192), Expect = 1e-14
Identities = 51/151 (33%), Positives = 84/151 (54%), Gaps = 4/151 (2%)
Frame = +3
Query: 618 IKASAKLTLLHSMLKILYKEGH--RVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVS 675
+++S K++ L +L + + ++FSQ K+L +LE+ L G KT R+DG+++
Sbjct: 189 VRSSTKVSTLIKLLTESRDQNPATKSVVFSQFRKMLLLLEEPLKAA-GFKTL-RLDGTMN 362
Query: 676 VTDRQTAIARFNQDK--SRFVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRA 733
R I +F + + L S R+ GINL A V + + +NP + QAM+R
Sbjct: 363 AKQRAQVIEQFQLSEVDEPMILLASLRASSTGINLTAASRVYLMEPWWNPAVEEQAMDRV 542
Query: 734 HRIGQSNRLLVYRLVVRASVEERILQLAKKK 764
HRIGQ + + RL+ + S+EE+IL L +KK
Sbjct: 543 HRIGQKEEVKIVRLIAKNSIEEKILMLQEKK 635
>TC91548 similar to PIR|D86185|D86185 hypothetical protein [imported] -
Arabidopsis thaliana, complete
Length = 611
Score = 78.2 bits (191), Expect = 1e-14
Identities = 41/109 (37%), Positives = 65/109 (59%)
Frame = +2
Query: 672 GSVSVTDRQTAIARFNQDKSRFVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMN 731
GS+++T R AI +F D +FL+S ++ G+ +NL A V + D +NP + QA +
Sbjct: 14 GSMTLTARDNAIKKFTDDPDCKIFLMSLKAGGVALNLTVASHVFLMDPWWNPAVERQAQD 193
Query: 732 RAHRIGQSNRLLVYRLVVRASVEERILQLAKKKLMLDQLFKGKSGSQKE 780
R HRIGQ + + R V+ ++EERIL+L +KK + +F+G G E
Sbjct: 194 RIHRIGQYKPIRIVRFVIENTIEERILKLQEKK---ELVFEGAVGGSSE 331
>BF632064 similar to PIR|H84432|H84 probable helicase [imported] -
Arabidopsis thaliana, partial (26%)
Length = 646
Score = 78.2 bits (191), Expect = 1e-14
Identities = 48/158 (30%), Positives = 81/158 (50%), Gaps = 12/158 (7%)
Frame = +3
Query: 471 RVLLTGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFNDLTSAEKVDELKKLVSPHMLR 530
R++LTGTPLQN+L E++++L F+ P F S ++ + +K ++ P +LR
Sbjct: 9 RLMLTGTPLQNDLHELWSMLEFMMPDIFASEDVDLKKLLGAEDKDLTSRMKSILGPFILR 188
Query: 531 RLKKDAMQNIPPKTERMVPVELSSIQAEYYR-----------AMLTKNYQI-LRNIGKGI 578
RLK D MQ + KT+++ V + Q Y+ A LTK + +N+ + +
Sbjct: 189 RLKSDVMQQLVRKTQKVQYVIMEKQQEHAYKEAIEEYRAVSQARLTKCSDLNPKNVLEVL 368
Query: 579 AQQSMLNIVMQLRKVCNHPYLIPGTEPDSGSVEFLHEM 616
++ + N +Q RK+ NHP LI D V F ++
Sbjct: 369 PRRQINNYFVQFRKIANHPLLIRRIYNDEDVVRFARKL 482
>BI311783 homologue to PIR|T47587|T4 TATA box binding protein (TBP)
associated factor (TAF)-like protein - Arabidopsis
thaliana, partial (6%)
Length = 788
Score = 73.9 bits (180), Expect = 3e-13
Identities = 40/121 (33%), Positives = 74/121 (61%), Gaps = 3/121 (2%)
Frame = +2
Query: 696 LLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEE 755
LL+T GLG+NL +ADT++ + D+NP D+QAM+RAHR+GQ + V+RL++R ++EE
Sbjct: 5 LLTTHVGGLGLNLTSADTLVFVEHDWNPMRDLQAMDRAHRLGQKKVVNVHRLIMRGTLEE 184
Query: 756 RILQLAKKKL-MLDQLFKGKSGSQK--EVEDILKWGTEELFNDSCALNGKDTSENNNSNK 812
+++ L + K+ + + + ++ S K + +L +LF + G ++++ N
Sbjct: 185 KVMSLQRFKVSVANAVINAENASLKTMNTDQLL-----DLFASAEIPKGSSVAKSSEDNS 349
Query: 813 D 813
D
Sbjct: 350 D 352
>TC84267 weakly similar to PIR|S41478|S41478 DNA repair protein rad8 -
fission yeast (Schizosaccharomyces pombe), partial (5%)
Length = 751
Score = 69.3 bits (168), Expect = 6e-12
Identities = 44/126 (34%), Positives = 70/126 (54%), Gaps = 1/126 (0%)
Frame = +2
Query: 640 RVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFNQDKSRFVFLLST 699
R +IFS T+ L ++E YL + R+DG S+ +RQ + +F + V +++T
Sbjct: 134 RSIIFSCWTRTLRLVERYLRA--AGIEHLRIDGDCSLPERQARLKQFADGDDKRVLIMTT 307
Query: 700 RSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVE-ERIL 758
+ G+NL A+ V I + +NP + QA+ RA R+GQ +LV R V+R +VE E
Sbjct: 308 GTGAFGLNLTCANRVFIVELQWNPSVENQAIARAIRLGQGREVLVTRYVMRDTVEVEMSS 487
Query: 759 QLAKKK 764
Q + KK
Sbjct: 488 QQSHKK 505
>CB065331 similar to PIR|D83545|D83 probable helicase PA0799 [imported] -
Pseudomonas aeruginosa (strain PAO1), partial (20%)
Length = 458
Score = 68.2 bits (165), Expect = 1e-11
Identities = 53/154 (34%), Positives = 76/154 (48%), Gaps = 5/154 (3%)
Frame = +1
Query: 506 ERFNDLTSAEKVDELKKLVSPHMLRRLKKDAMQNIPPKTERMVPVELSSIQAEYY---RA 562
ER D AE++ L + P +LRR K+ +P KTE + VELS Q + Y R
Sbjct: 34 ERHGD---AERMAHLAARIRPFLLRRTKEQVATELPAKTEMVHWVELSDAQRDTYETVRV 204
Query: 563 MLTKNY--QILRNIGKGIAQQSMLNIVMQLRKVCNHPYLIPGTEPDSGSVEFLHEMRIKA 620
+ K +I RN G +Q +L+ +++LR+VC L+ G EP +
Sbjct: 205 AMDKKVRDEIARN-GAARSQIVILDALLKLRQVCCDLRLVKGIEPKGSQAD--------- 354
Query: 621 SAKLTLLHSMLKILYKEGHRVLIFSQMTKLLDIL 654
KL L ML L EG RVL+FSQ T +L ++
Sbjct: 355 KGKLGALLDMLDELLXEGRRVLLFSQFTSMLALI 456
>TC83036 similar to PIR|C96637|C96637 hypothetical protein F11P17.13
[imported] - Arabidopsis thaliana, partial (20%)
Length = 1256
Score = 62.0 bits (149), Expect = 1e-09
Identities = 41/174 (23%), Positives = 85/174 (48%), Gaps = 12/174 (6%)
Frame = +2
Query: 439 VPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLLTGTPLQNNLGEMYNLLNFLQPASF 498
V W +++DE +KN +++ + + R L+GTP+QN + ++Y+ FL+ +
Sbjct: 53 VAWFRVVLDEAQSIKNHRTQVARACWGLRAKRRWCLSGTPIQNAIDDLYSYFRFLRYDPY 232
Query: 499 PSLSAFEERFN---DLTSAEKVDELKKLVSPHMLRRLKKDAMQ-----NIPPKTERMVPV 550
++F + ++ +L+ ++ MLRR K + ++PPK+ + V
Sbjct: 233 AVYTSFCSTIKIPINRNPSKGYRKLQAVLKTIMLRRTKGTLLDGEPIISLPPKSVELRKV 412
Query: 551 ELSSIQAEYYRAM-LTKNYQILRNIGKGIAQQSMLNI---VMQLRKVCNHPYLI 600
E S + ++Y + Q G +Q+ +NI +++LR+ C+HP L+
Sbjct: 413 EFSQEERDFYSKLEADSRAQFQEYADAGTVKQNYVNILLMLLRLRQACDHPLLV 574
>BQ122366 similar to PIR|T47325|T4 hypothetical protein T12K4.120 -
Arabidopsis thaliana, partial (10%)
Length = 462
Score = 58.9 bits (141), Expect = 9e-09
Identities = 37/103 (35%), Positives = 57/103 (54%), Gaps = 1/103 (0%)
Frame = -3
Query: 694 VFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASV 753
+ LLS +C GI+L A VI DS++NP QA+ RA R GQ + VY L++ S+
Sbjct: 421 ILLLSINACAEGISLTAASRVIFLDSEWNPSKTKQAIARAFRPGQEKMVYVYHLLMTGSM 242
Query: 754 EE-RILQLAKKKLMLDQLFKGKSGSQKEVEDILKWGTEELFND 795
EE + + K+ + +F S++ VED KW E++ +D
Sbjct: 241 EEDKYRRTTWKEWVSCMIF-----SEELVEDPSKWQAEKIEDD 128
>TC87517 similar to PIR|F84833|F84833 probable SNF2/SWI2 family
transcription factor [imported] - Arabidopsis thaliana,
partial (6%)
Length = 773
Score = 54.7 bits (130), Expect = 2e-07
Identities = 27/71 (38%), Positives = 44/71 (61%)
Frame = +2
Query: 694 VFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASV 753
V LL + G+NL A V++ + NP A+ QA++R HRIGQ + L++R +V+ +V
Sbjct: 143 VLLLLIQHGANGLNLLEAQHVVLVEPLLNPAAEAQAISRVHRIGQKQKTLIHRFLVKDTV 322
Query: 754 EERILQLAKKK 764
EE I +L + +
Sbjct: 323 EESIYKLNRSR 355
>BQ751767 similar to SP|O10302|GTA_N Probable global transactivator. [OpMNPV]
{Orgyia pseudotsugata multicapsid polyhedrosis virus},
partial (7%)
Length = 771
Score = 52.8 bits (125), Expect = 6e-07
Identities = 47/203 (23%), Positives = 85/203 (41%), Gaps = 27/203 (13%)
Frame = +1
Query: 329 ILADEMGLGKTISACAFISSLYFEFKV-----------------SRPCLVLVP-LVTMGN 370
ILADEMG+GK++S + I +V S LV+V + + N
Sbjct: 73 ILADEMGMGKSLSILSLIVKTLDSGRVWAEEESNSEERSKDMAYSGATLVVVSSALLISN 252
Query: 371 WLAEFALWAPDVNVV-----QYHGCAKARAIIRQYEWHASDPSGLNKKTEAYKFNVLLTS 425
W AE A + C + ++H P + + K ++++T+
Sbjct: 253 WEAEVDK*ASPTMPI*RPTESLTLCRHVEGGLTYVKYHG--PGRETEMDKIKKTDMVVTT 426
Query: 426 YEMVLADY----SHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLLTGTPLQN 481
Y + A++ S + W +++DE H ++ + + ++ R LTGTP+QN
Sbjct: 427 YNTLSAEFDRKESRLHKIGWYRVVLDEAHIIRRPATTFYQACRALQANSRWCLTGTPIQN 606
Query: 482 NLGEMYNLLNFLQPASFPSLSAF 504
L ++ LL F++ + F F
Sbjct: 607 RLSDIGALLAFIRASPFDDPRVF 675
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.314 0.131 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,044,662
Number of Sequences: 36976
Number of extensions: 361820
Number of successful extensions: 1449
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 1407
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1427
length of query: 935
length of database: 9,014,727
effective HSP length: 105
effective length of query: 830
effective length of database: 5,132,247
effective search space: 4259765010
effective search space used: 4259765010
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146651.1


