

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146650.9 - phase: 0 /pseudo
(314 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
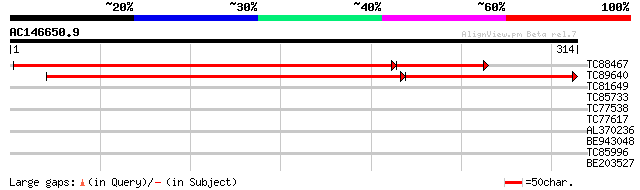
TC88467 similar to GP|16648927|gb|AAL24315.1 formyltetrahydrofol... 435 e-146
TC89640 similar to GP|16648927|gb|AAL24315.1 formyltetrahydrofol... 297 e-103
TC81649 similar to GP|10178116|dbj|BAB11409. ubiquitin carboxyl-... 30 0.92
TC85733 similar to GP|3832512|gb|AAC70779.1| granule-bound glyco... 28 4.6
TC77538 similar to SP|P54237|G6P1_CLAMI Glucose-6-phosphate isom... 28 4.6
TC77617 similar to GP|6671947|gb|AAF23207.1| putative ubiquitin ... 28 6.0
AL370236 weakly similar to GP|6041804|gb| putative protein kinas... 28 6.0
BE943048 similar to GP|16930515|gb| AT5g44240/MLN1_17 {Arabidops... 28 6.0
TC85996 weakly similar to GP|16648871|gb|AAL24287.1 Unknown prot... 27 7.8
BE203527 homologue to GP|7705248|gb| plasma membrane H(+)-ATPase... 27 7.8
>TC88467 similar to GP|16648927|gb|AAL24315.1 formyltetrahydrofolate
deformylase-like {Arabidopsis thaliana}, partial (70%)
Length = 836
Score = 435 bits (1118), Expect(2) = e-146
Identities = 212/212 (100%), Positives = 212/212 (100%)
Frame = +3
Query: 3 MVVRRVSQVLGLTNSNKIRNFSFKSLDLPPLPSPSLSHGIHVFHCPDAVGIVAKLSECIA 62
MVVRRVSQVLGLTNSNKIRNFSFKSLDLPPLPSPSLSHGIHVFHCPDAVGIVAKLSECIA
Sbjct: 3 MVVRRVSQVLGLTNSNKIRNFSFKSLDLPPLPSPSLSHGIHVFHCPDAVGIVAKLSECIA 182
Query: 63 SRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQMEEDFLKLSQAFNATRSVVRVPA 122
SRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQMEEDFLKLSQAFNATRSVVRVPA
Sbjct: 183 SRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQMEEDFLKLSQAFNATRSVVRVPA 362
Query: 123 LDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVISNHHRDSNTHVIRFLERHGIPY 182
LDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVISNHHRDSNTHVIRFLERHGIPY
Sbjct: 363 LDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVISNHHRDSNTHVIRFLERHGIPY 542
Query: 183 HCLSTTNENKREGEILELVQNTDFLVLARYMQ 214
HCLSTTNENKREGEILELVQNTDFLVLARYMQ
Sbjct: 543 HCLSTTNENKREGEILELVQNTDFLVLARYMQ 638
Score = 100 bits (249), Expect(2) = e-146
Identities = 51/51 (100%), Positives = 51/51 (100%)
Frame = +1
Query: 215 ATGMT*LTFTMVCCHHSRVVIHLNRLLRLV*N*LAQQVTLCPKNLTADQ*L 265
ATGMT*LTFTMVCCHHSRVVIHLNRLLRLV*N*LAQQVTLCPKNLTADQ*L
Sbjct: 664 ATGMT*LTFTMVCCHHSRVVIHLNRLLRLV*N*LAQQVTLCPKNLTADQ*L 816
>TC89640 similar to GP|16648927|gb|AAL24315.1 formyltetrahydrofolate
deformylase-like {Arabidopsis thaliana}, partial (83%)
Length = 1126
Score = 297 bits (761), Expect(2) = e-103
Identities = 141/199 (70%), Positives = 165/199 (82%)
Frame = +1
Query: 21 RNFSFKSLDLPPLPSPSLSHGIHVFHCPDAVGIVAKLSECIASRGGNILAADVFVPQNKH 80
RN+S S + S L+HGIHVF CPD++GIVAKLS+CIAS GGNI+AADVFVPQNK
Sbjct: 145 RNYSHNSFNDNSASSSFLTHGIHVFQCPDSIGIVAKLSDCIASNGGNIIAADVFVPQNKG 324
Query: 81 VFYSRSDFVFDPVKWPRKQMEEDFLKLSQAFNATRSVVRVPALDPKYKIAVLASKQDHCL 140
+FYSR+DFVFD VKWPR +MEEDFLK+S+ +NA RS+++VPALDPKYKIAVLAS QDHCL
Sbjct: 325 LFYSRTDFVFDHVKWPRLRMEEDFLKISKTYNAVRSILKVPALDPKYKIAVLASNQDHCL 504
Query: 141 VDLLHGWQDGKLPVDITCVISNHHRDSNTHVIRFLERHGIPYHCLSTTNENKREGEILEL 200
+D LHGWQDG+LPVDITCVISNH R + VIRFL+RH IPYH L TT ENKRE +IL+L
Sbjct: 505 IDSLHGWQDGRLPVDITCVISNHDRGPESEVIRFLQRHNIPYHYLKTTKENKREDDILKL 684
Query: 201 VQNTDFLVLARYMQATGMT 219
VQ+TDFLVLARY + T
Sbjct: 685 VQDTDFLVLARYTKIISST 741
Score = 97.1 bits (240), Expect(2) = e-103
Identities = 52/95 (54%), Positives = 63/95 (65%)
Frame = +2
Query: 220 *LTFTMVCCHHSRVVIHLNRLLRLV*N*LAQQVTLCPKNLTADQ*LNKWLREFHTEMTCR 279
*LTFT+ CCHH IHL+R V* *L QQV L PK++ LNKWL+EF +MTCR
Sbjct: 767 *LTFTIACCHHLEAPIHLSRPSMQV*K*LVQQVIL*PKDVMLGLLLNKWLKEFLIKMTCR 946
Query: 280 ALCRNQKTLRNNVFPRLLDPIVNFGYYLMRLRRLL 314
LCRN++TLRNNV L D +F YY ++ R LL
Sbjct: 947 GLCRNRRTLRNNVCQWLSDLTASFEYYHIKKRILL 1051
>TC81649 similar to GP|10178116|dbj|BAB11409. ubiquitin carboxyl-terminal
hydrolase {Arabidopsis thaliana}, partial (24%)
Length = 905
Score = 30.4 bits (67), Expect = 0.92
Identities = 17/39 (43%), Positives = 20/39 (50%)
Frame = +1
Query: 4 VVRRVSQVLGLTNSNKIRNFSFKSLDLPPLPSPSLSHGI 42
VV RVSQ LGL + +KIR S P P P G+
Sbjct: 175 VVERVSQHLGLNDPSKIRLTSHNCYSQQPKPQPIKYRGV 291
>TC85733 similar to GP|3832512|gb|AAC70779.1| granule-bound glycogen (starch)
synthase {Astragalus membranaceus}, partial (94%)
Length = 2251
Score = 28.1 bits (61), Expect = 4.6
Identities = 13/31 (41%), Positives = 17/31 (53%)
Frame = -1
Query: 161 SNHHRDSNTHVIRFLERHGIPYHCLSTTNEN 191
S HHRD H R + +HGI Y LS ++
Sbjct: 1648 SLHHRDQQFHTPR*MLQHGILYSLLSQCQQD 1556
>TC77538 similar to SP|P54237|G6P1_CLAMI Glucose-6-phosphate isomerase
cytosolic 1 (EC 5.3.1.9) (GPI) (Phosphoglucose
isomerase) (PGI), complete
Length = 2215
Score = 28.1 bits (61), Expect = 4.6
Identities = 12/20 (60%), Positives = 15/20 (75%)
Frame = +3
Query: 22 NFSFKSLDLPPLPSPSLSHG 41
+ S+ S LPPL SP+LSHG
Sbjct: 180 SISYSSKWLPPLSSPTLSHG 239
>TC77617 similar to GP|6671947|gb|AAF23207.1| putative ubiquitin
carboxyl-terminal hydrolase {Arabidopsis thaliana},
partial (27%)
Length = 1473
Score = 27.7 bits (60), Expect = 6.0
Identities = 15/39 (38%), Positives = 19/39 (48%)
Frame = +1
Query: 4 VVRRVSQVLGLTNSNKIRNFSFKSLDLPPLPSPSLSHGI 42
VV RV+Q LGL + +KIR P P P G+
Sbjct: 82 VVERVAQQLGLDDPSKIRLTPHNCYSQQPKPQPIKHRGV 198
>AL370236 weakly similar to GP|6041804|gb| putative protein kinase
{Arabidopsis thaliana}, partial (4%)
Length = 378
Score = 27.7 bits (60), Expect = 6.0
Identities = 11/30 (36%), Positives = 17/30 (56%)
Frame = -3
Query: 212 YMQATGMT*LTFTMVCCHHSRVVIHLNRLL 241
Y+ + TF +CC H R+VI L+ L+
Sbjct: 361 YLNSAKTIIATFFNICCSHGRIVIKLSLLI 272
>BE943048 similar to GP|16930515|gb| AT5g44240/MLN1_17 {Arabidopsis
thaliana}, partial (20%)
Length = 503
Score = 27.7 bits (60), Expect = 6.0
Identities = 13/36 (36%), Positives = 19/36 (52%)
Frame = +1
Query: 178 HGIPYHCLSTTNENKREGEILELVQNTDFLVLARYM 213
H YHCL+TT E R +I + ++ L L Y+
Sbjct: 250 HSQDYHCLTTTQEIARTSKINKQLEEQHMLQLNLYI 357
>TC85996 weakly similar to GP|16648871|gb|AAL24287.1 Unknown protein
{Arabidopsis thaliana}, partial (53%)
Length = 1446
Score = 27.3 bits (59), Expect = 7.8
Identities = 16/64 (25%), Positives = 26/64 (40%), Gaps = 10/64 (15%)
Frame = -3
Query: 160 ISNHHRDSNTHVIRFLERHGIPYHCL----------STTNENKREGEILELVQNTDFLVL 209
I HHR+ TH F H I +T N N++ + +++ QN L+
Sbjct: 1387 IVQHHREHPTHTYTFTLNHAITIKSTTHSTLKNNKPNTMNTNRKSPQHIDIYQN*TSLLS 1208
Query: 210 ARYM 213
Y+
Sbjct: 1207 VNYL 1196
>BE203527 homologue to GP|7705248|gb| plasma membrane H(+)-ATPase precursor
{Vicia faba}, partial (14%)
Length = 417
Score = 27.3 bits (59), Expect = 7.8
Identities = 14/49 (28%), Positives = 24/49 (48%), Gaps = 2/49 (4%)
Frame = -1
Query: 145 HGWQDGKLPVDITCVISNHHRDSNTHVIRFLERHGI--PYHCLSTTNEN 191
H W++ KLP + H+ DS+ H I + H + C +T++N
Sbjct: 147 HEWREIKLPYQCNKHEAKHYTDSDRHCINRIVLHSLEDSSTC*DSTDDN 1
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.333 0.144 0.459
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,316,501
Number of Sequences: 36976
Number of extensions: 170378
Number of successful extensions: 1579
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 1566
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1579
length of query: 314
length of database: 9,014,727
effective HSP length: 96
effective length of query: 218
effective length of database: 5,465,031
effective search space: 1191376758
effective search space used: 1191376758
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146650.9


