

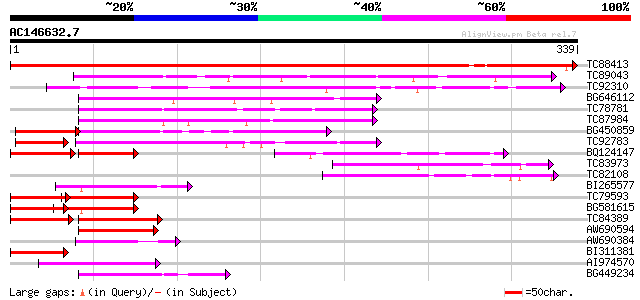
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146632.7 + phase: 0
(339 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC88413 weakly similar to GP|10176786|dbj|BAB09900. gene_id:MIK1... 605 e-174
TC89043 similar to GP|10178241|dbj|BAB11673. gene_id:MDJ22.9~unk... 119 1e-27
TC92310 weakly similar to PIR|T47778|T47778 hypothetical protein... 75 3e-14
BG646112 weakly similar to GP|10177368|dbj contains similarity t... 69 3e-12
TC78781 weakly similar to PIR|T02291|T02291 hypothetical protein... 65 5e-11
TC87984 weakly similar to GP|10177368|dbj|BAB10659. contains sim... 62 2e-10
BG450859 weakly similar to GP|21553400|gb| unknown {Arabidopsis ... 54 9e-08
TC92783 similar to PIR|T49020|T49020 hypothetical protein F3C22.... 52 3e-07
BQ124147 similar to GP|10177836|dbj emb|CAB62440.1~gene_id:MCD7.... 52 3e-07
TC83973 GP|21537320|gb|AAM61661.1 unknown {Arabidopsis thaliana}... 52 4e-07
TC82108 similar to GP|21537320|gb|AAM61661.1 unknown {Arabidopsi... 50 2e-06
BI265577 similar to GP|10177368|dbj contains similarity to heat ... 50 2e-06
TC79593 similar to GP|10177368|dbj|BAB10659. contains similarity... 47 1e-05
BG581615 similar to GP|10177368|dbj contains similarity to heat ... 46 2e-05
TC84389 weakly similar to GP|10177368|dbj|BAB10659. contains sim... 45 4e-05
AW690594 similar to GP|23498163|emb hypothetical protein {Plasmo... 45 5e-05
AW690384 similar to GP|23496006|gb hypothetical protein {Plasmod... 45 5e-05
BI311381 weakly similar to PIR|T46128|T461 hypothetical protein ... 44 7e-05
AI974570 similar to GP|10177368|dbj contains similarity to heat ... 44 1e-04
BG449234 weakly similar to GP|8979942|gb|A Contains a F-box PF|0... 42 4e-04
>TC88413 weakly similar to GP|10176786|dbj|BAB09900.
gene_id:MIK19.29~pir||T01344~similar to unknown protein
{Arabidopsis thaliana}, partial (10%)
Length = 1288
Score = 605 bits (1561), Expect = e-174
Identities = 308/343 (89%), Positives = 318/343 (91%), Gaps = 4/343 (1%)
Frame = +1
Query: 1 MSGLPDELLCEILSFLPSEEFPSTSLLSKRWRRVWLGMPNADRISILPDELLCRILSLLP 60
MSGLPDELLC+ILSFLPSEE PSTSLLSKRWRRVWLGMPNADRISILPDELLCRILSLLP
Sbjct: 124 MSGLPDELLCQILSFLPSEEIPSTSLLSKRWRRVWLGMPNADRISILPDELLCRILSLLP 303
Query: 61 AKQIMVTSLLSRRWRSLRPRMTEINIDDTSYIHDRDAYDRYYHVIALFLFEMKIHHPIMK 120
AKQIMVTSLLSRRWRSLRPRMTEINIDDTSYIHDRDAYDRYYHVIALFLFEMKIHHPIMK
Sbjct: 304 AKQIMVTSLLSRRWRSLRPRMTEINIDDTSYIHDRDAYDRYYHVIALFLFEMKIHHPIMK 483
Query: 121 TVTILSASPFFNPLPLWLGCLQVQHLDVTSSATLCLCVPYKVLNSTALVVLKLNALTIDY 180
TVTILSASPFFNPLPLWL CL+VQHLDVTSSATLCLCVPYKVL STALVVLKLNALTIDY
Sbjct: 484 TVTILSASPFFNPLPLWLNCLKVQHLDVTSSATLCLCVPYKVLTSTALVVLKLNALTIDY 663
Query: 181 VHRSSTNLPSLKILHLTQVHFLKLKFLIKILSMSPLLEDLLLKDLQVTDNTLAHDDAAAL 240
VHRSSTNLPSLKILHLTQVHFLKLKFLIKILSMSPLLEDLLLKDLQVTDNTLA DDAAAL
Sbjct: 664 VHRSSTNLPSLKILHLTQVHFLKLKFLIKILSMSPLLEDLLLKDLQVTDNTLAQDDAAAL 843
Query: 241 KPFPKLLRADVSESSISAFLLPLKLFYNVQFLRSQVLLQTLEQDFSTTQFLNLTHMDLIF 300
KPFPKLLRAD+S+S IS LLPLKLFYNV FLRSQ LQTLE+ TQFL+LTH+DL F
Sbjct: 844 KPFPKLLRADISDSCISPLLLPLKLFYNVHFLRSQ--LQTLEEQ-QDTQFLSLTHLDLSF 1014
Query: 301 HDGYYWISLMKFICACPSLQTLTIHNIGGDY----DDHNNNWP 339
GYYWISL+KFICACPSLQTLTI IGG Y +D +NNWP
Sbjct: 1015DHGYYWISLIKFICACPSLQTLTIRKIGGGYGLLSNDDHNNWP 1143
>TC89043 similar to GP|10178241|dbj|BAB11673. gene_id:MDJ22.9~unknown
protein {Arabidopsis thaliana}, partial (9%)
Length = 1413
Score = 119 bits (299), Expect = 1e-27
Identities = 92/308 (29%), Positives = 152/308 (48%), Gaps = 19/308 (6%)
Frame = +2
Query: 39 PNADRISILPDELLCRILSLLPAKQIMVTSLLSRRWRSLRPRMTEINIDDTSYIHDRDAY 98
P DRIS LPD++LCRILS LP K T++LS+RW L +T ++ DD S + D D +
Sbjct: 53 PPVDRISHLPDDILCRILSFLPTKLAFTTTVLSKRWTPLYKLLTSLSFDDESVL-DEDTF 229
Query: 99 DRYYHVIALFLFEMKIHHPIMKTVTILSASP---FFNPLPLWLGCLQVQHLDVTSSATLC 155
R+ + F ++KT+ + SP FN L LW+G + ++ +
Sbjct: 230 LRFCRFVDTVTFST----DLIKTLHLNCGSPNWKHFN-LDLWIGTAKRHPVENFNLVGTW 394
Query: 156 LCVPYK--VLNSTALVVLKLNALTIDYVHRSSTNLPSLKILHLTQVHFLKLKFLIKILSM 213
+P + + +LVVLKL L I V + +LP LKILHL +V+ KIL
Sbjct: 395 RSIPLRPSIFRFPSLVVLKLKTLKI-IVGNITVDLPLLKILHLDRVYLKNKTNFNKILYG 571
Query: 214 SPLLEDLLLKDLQVTDNTLAHDDAAAL---------KPFPKLLRADVSESSISAFLLPLK 264
P+LED L+ ++ + T D+ L K PKL+R ++ + P +
Sbjct: 572 CPVLED-LIANIYYKEPTPEPDEVFTLSKATATGEFKILPKLIRVQINADEV-----PFR 733
Query: 265 LFYNVQFLRSQVLLQTLEQDFSTTQ-----FLNLTHMDLIFHDGYYWISLMKFICACPSL 319
+NV+FL + + + + ++ F NL + L ++ ++W +M+ + CP++
Sbjct: 734 AIHNVEFLALTMRSRLPDPEINSYNILSPIFRNLILLQLCMYNFHHWDHVMEVLQHCPNI 913
Query: 320 QTLTIHNI 327
Q L I+ +
Sbjct: 914 QVLRINKL 937
>TC92310 weakly similar to PIR|T47778|T47778 hypothetical protein F17J16.10
- Arabidopsis thaliana, partial (8%)
Length = 1019
Score = 75.5 bits (184), Expect = 3e-14
Identities = 86/317 (27%), Positives = 130/317 (40%), Gaps = 7/317 (2%)
Frame = +2
Query: 23 STSLLSKRWRRVWLGMPNADRISILPDELLCRILSLLPAKQIMVTSLLSRRWRSLRPRMT 82
+TS+LSKRW +W +P D D L S L Q T +L R +
Sbjct: 110 TTSVLSKRWTNLWCFVPFLD----FSDIKLADHESFLWFNQFFCTVMLFRETYAS----- 262
Query: 83 EINIDDTSYIHDRDAYDRYYHVIALFLFEMKIHHPIMKTVTILSASPFFNPLPLWLGCLQ 142
N D+ + + A+ Y+ ++ L L L L L L
Sbjct: 263 --NSIDSFSLDIQYAHLNYFSILRL------------NNWLYLLGKRNVKYLNLHLNFLN 400
Query: 143 VQHLDVTSSATLCLCVPYKVLNSTALVVLKLNALTIDYVHRSSTNL----PSLKILHLTQ 198
+DV TL +P + LVVL ++ SS PSLK LH
Sbjct: 401 ALLVDVQHRKTLTPKLPSTIFTCKTLVVLNISWFAFKGFSFSSVGFGFGFPSLKTLHFNH 580
Query: 199 VHFLKLKFLIKILSMSPLLEDLLLKDLQVTDNTLAHDDAAALKP---FPKLLRADVSESS 255
++F L + +L+ P+LED K V TL ++ A + L+R D+ +++
Sbjct: 581 IYFNHLSDFLLLLAGCPVLED--FKACHVF--TLEEEEEATQESTLNLSNLIRTDIIDTN 748
Query: 256 ISAFLLPLKLFYNVQFLRSQVLLQTLEQDFSTTQFLNLTHMDLIFHDGYYWISLMKFICA 315
F +P+K +N FLR Q+ + DF T F NLTH+ + YY +K +
Sbjct: 749 ---FDIPMKALFNSVFLRIQLCQRYTSYDFPT--FNNLTHLVI----NYYLDMFIKVLYH 901
Query: 316 CPSLQTLTIHNIGGDYD 332
CP+LQ L ++ D D
Sbjct: 902 CPNLQKLELYQTQTDCD 952
>BG646112 weakly similar to GP|10177368|dbj contains similarity to heat shock
transcription factor HSF30~gene_id:K16L22.13, partial
(5%)
Length = 742
Score = 68.9 bits (167), Expect = 3e-12
Identities = 63/193 (32%), Positives = 93/193 (47%), Gaps = 12/193 (6%)
Frame = +2
Query: 42 DRISILPDELLCRILSLLPAK-QIMVTSLLSRRWRSLRPRMTEINIDDTSYIHDRD--AY 98
DR+S LPD LLC ILS LP K +M TSL+SRRWR L + DD S + R+ +
Sbjct: 14 DRMSSLPDSLLCHILSFLPTKTSVMTTSLVSRRWRHLWEHLNVFVFDDKSNCNCRNPKKF 193
Query: 99 DRYYHVIALFLFEMKIHHPIMKTVTILSASPFFNP---LPLWLGCLQVQHL-DVTSSATL 154
++ ++ L K H +T ++ + P + +W+ HL D++ + T
Sbjct: 194 RKFAFFVSSVLSLRKSRHIRKFHLTCCTSDVYSFPGECVYMWVRAAIGPHLEDLSLNITN 373
Query: 155 C-----LCVPYKVLNSTALVVLKLNALTIDYVHRSSTNLPSLKILHLTQVHFLKLKFLIK 209
C + +P +LN T LV L L L S+ + PSLK+L +V F L+ I
Sbjct: 374 CHGDDMVYLPPSLLNCTNLVSLSLFGLIHLKFQPSAIHFPSLKML---KVEFSILEHNID 544
Query: 210 ILSMSPLLEDLLL 222
I L D +L
Sbjct: 545 IEDFGKHLTDSIL 583
>TC78781 weakly similar to PIR|T02291|T02291 hypothetical protein T13D8.28 -
Arabidopsis thaliana, partial (12%)
Length = 1300
Score = 64.7 bits (156), Expect = 5e-11
Identities = 60/180 (33%), Positives = 91/180 (50%), Gaps = 1/180 (0%)
Frame = +3
Query: 42 DRISILPDELLCRILSLLPAKQIMVTSLLSRRWRSLRPRMTEINIDDTSYIHDRDAYDRY 101
DRIS L D+LL RILS L ++ + T +LS+RW + +T + +DD + DA D
Sbjct: 129 DRISSLSDDLLVRILSNLHTRESVSTCVLSKRWVHVFKALTCLRLDDENG-SLLDALDEM 305
Query: 102 YHVIALFLFEMKIHHPIMKTVTILSASPFFNPLPLWLGCLQVQHLDVTSSATLCLCVPYK 161
H L F++ I + ++ N + L L + L + + L P
Sbjct: 306 RHRTDLTSFDLSIFTRRSGDIVTVAEKAVRNIVRL---NLSLNSLSIYGILNMYLTRP-- 470
Query: 162 VLNSTALVVLKLNALTIDYVHRSSTNLPSLKILHLTQVHF-LKLKFLIKILSMSPLLEDL 220
V NS LV LKL+ + I S +LPSLK+L L++V F K FL + ++S +LE+L
Sbjct: 471 VFNSRTLVELKLHRVCIAET-LSVASLPSLKVLSLSRVQFDTKSVFLSLLSAVSRVLEEL 647
>TC87984 weakly similar to GP|10177368|dbj|BAB10659. contains similarity to
heat shock transcription factor HSF30~gene_id:K16L22.13,
partial (5%)
Length = 696
Score = 62.4 bits (150), Expect = 2e-10
Identities = 59/195 (30%), Positives = 93/195 (47%), Gaps = 16/195 (8%)
Frame = +3
Query: 42 DRISILPDELLCRILSLLPAKQ-IMVTSLLSRRWRSLRPRMTEINIDDTS----YIHDRD 96
DRI+ LP+ LL ILS LP K + T L+SR+WR+L + ++ D+S Y D D
Sbjct: 105 DRINQLPNSLLHHILSFLPTKTCVQTTPLVSRKWRNLWKNLEALDFCDSSSPQIYEFDND 284
Query: 97 AYDRYYHVI---ALFLFEMKIHHPIMKTVTILSASPFFN-PLPLWLGC-----LQVQHLD 147
+ V L L + ++ + + PF+N + W+ L+ HL
Sbjct: 285 EQFLIFSVFVNTVLTLRKSRVVRKFCLSCYHVQLDPFYNHSIDTWINATIGPNLEEFHLT 464
Query: 148 VTSSATLCLCVPYKVLNSTALVVLKLN-ALTIDYVHRSSTNLPSLKILHLTQVHFLKLKF 206
+ ++A VP + + LV L N + + S LPSLK+L L ++ L LKF
Sbjct: 465 LLTAAGFNR-VPLSLFSCPNLVSLSFNDYIILQLQDNSKICLPSLKLLQLLDMYNLDLKF 641
Query: 207 LIKILSMS-PLLEDL 220
S++ P+LE+L
Sbjct: 642 CEMPFSLAVPVLENL 686
>BG450859 weakly similar to GP|21553400|gb| unknown {Arabidopsis thaliana},
partial (11%)
Length = 549
Score = 53.9 bits (128), Expect = 9e-08
Identities = 49/153 (32%), Positives = 76/153 (49%), Gaps = 2/153 (1%)
Frame = +2
Query: 42 DRISILPDELLCRILSLLPAKQIMVTSLLSRRWRSLRPRMTEINIDDTSYIHDRDAYDRY 101
DRI LPDE+L ILS +P KQ TS+LS+RW L + ++ +T+ + DRD+ R+
Sbjct: 122 DRIGTLPDEILIHILSFVPTKQAFTTSILSKRWIHLWRYVPILDFTETN-LEDRDSVIRF 298
Query: 102 YHVIALFLFE-MKIHHPIMKTVTILSASPFFNPLPLWLGCLQVQHLDVTSSATLCLCVPY 160
F+F ++ H + S + F L + ++ L +S T+ P
Sbjct: 299 EE----FIFSVIRSRH----SAGNHSINTFI--LDIQRHSSRLHALVFGNSNTIAPEFPI 448
Query: 161 KVLNSTALVVLKLNALTIDY-VHRSSTNLPSLK 192
+L ST LV LKL + + + + LPSLK
Sbjct: 449 PILGSTTLVXLKLASFDMGADLFLNLITLPSLK 547
Score = 45.4 bits (106), Expect = 3e-05
Identities = 20/39 (51%), Positives = 29/39 (74%)
Frame = +2
Query: 4 LPDELLCEILSFLPSEEFPSTSLLSKRWRRVWLGMPNAD 42
LPDE+L ILSF+P+++ +TS+LSKRW +W +P D
Sbjct: 137 LPDEILIHILSFVPTKQAFTTSILSKRWIHLWRYVPILD 253
>TC92783 similar to PIR|T49020|T49020 hypothetical protein F3C22.70 -
Arabidopsis thaliana (fragment), partial (7%)
Length = 679
Score = 52.0 bits (123), Expect = 3e-07
Identities = 65/204 (31%), Positives = 88/204 (42%), Gaps = 21/204 (10%)
Frame = +1
Query: 40 NADRISILPDELLCRILSLLPAKQIMVT-SLLSRRWRSLRPRMTEINIDDTSYIHDRDAY 98
N D I+ LPD LLC ILS LP K + T L+SRRWR L + + H +
Sbjct: 88 NTDWINTLPDSLLCHILSFLPTKITVTTIPLVSRRWRHLWENLQVFDFYLKYNKHTINNN 267
Query: 99 D-RYYHVIALFLFEMKIHHPIMKTVTILSAS---PFF-NPLPLWL-----GCLQVQHLDV 148
D R + + ++ I K S PFF N + W+ CL Q LD+
Sbjct: 268 DFRKFAAFVNSVLSIRRSRDIRKMRLSCGHSEVDPFFSNSIDSWIRSATGSCL--QELDI 441
Query: 149 T---------SSATLCLCVPYKVLNSTALVVLKLNALTIDYVHRSST-NLPSLKILHLTQ 198
T S L + P T LV L L + RSS LPSLK L L
Sbjct: 442 TLFYTDREYVFSIFLTILAP-----CTNLVSLSLCGDIYVVLERSSAFFLPSLKKLQL-D 603
Query: 199 VHFLKLKFLIKILSMSPLLEDLLL 222
+ ++++ + +LS P+LE L L
Sbjct: 604 IGYVEMNSMDNLLSSCPILETLEL 675
Score = 41.2 bits (95), Expect = 6e-04
Identities = 19/33 (57%), Positives = 25/33 (75%), Gaps = 1/33 (3%)
Frame = +1
Query: 4 LPDELLCEILSFLPSEEFPST-SLLSKRWRRVW 35
LPD LLC ILSFLP++ +T L+S+RWR +W
Sbjct: 109 LPDSLLCHILSFLPTKITVTTIPLVSRRWRHLW 207
>BQ124147 similar to GP|10177836|dbj emb|CAB62440.1~gene_id:MCD7.13~similar
to unknown protein {Arabidopsis thaliana}, partial
(11%)
Length = 677
Score = 52.0 bits (123), Expect = 3e-07
Identities = 24/36 (66%), Positives = 30/36 (82%)
Frame = +1
Query: 42 DRISILPDELLCRILSLLPAKQIMVTSLLSRRWRSL 77
DRISILPD++L ILS LP KQ VTS+LS+RW++L
Sbjct: 25 DRISILPDDILVHILSPLPTKQAFVTSVLSKRWKNL 132
Score = 50.4 bits (119), Expect = 1e-06
Identities = 47/145 (32%), Positives = 67/145 (45%), Gaps = 5/145 (3%)
Frame = +1
Query: 159 PYKVLNSTALVVLKLNALTI-----DYVHRSSTNLPSLKILHLTQVHFLKLKFLIKILSM 213
P + LV+LKL+ L + +Y+ R LPSLK LHL + F + L +L
Sbjct: 259 PNTIFTRETLVILKLSELFMGSGFCNYLIR----LPSLKTLHLKDIEFQQYGDLDCLLEG 426
Query: 214 SPLLEDLLLKDLQVTDNTLAHDDAAALKPFPKLLRADVSESSISAFLLPLKLFYNVQFLR 273
P+LEDL L D+ ++ A K KL RAD+ + + +K NV+FL
Sbjct: 427 CPVLEDLQLYDISYVSLLFSN---ACRKTLTKLNRADIIQCDCG---VRMKALSNVEFLH 588
Query: 274 SQVLLQTLEQDFSTTQFLNLTHMDL 298
++ L F F NLTH L
Sbjct: 589 IKLSKGYLHYVFPA--FNNLTHFVL 657
Score = 41.6 bits (96), Expect = 4e-04
Identities = 19/39 (48%), Positives = 29/39 (73%)
Frame = +1
Query: 1 MSGLPDELLCEILSFLPSEEFPSTSLLSKRWRRVWLGMP 39
+S LPD++L ILS LP+++ TS+LSKRW+ +W +P
Sbjct: 31 ISILPDDILVHILSPLPTKQAFVTSVLSKRWKNLWCLVP 147
>TC83973 GP|21537320|gb|AAM61661.1 unknown {Arabidopsis thaliana}, partial
(2%)
Length = 870
Score = 51.6 bits (122), Expect = 4e-07
Identities = 46/141 (32%), Positives = 72/141 (50%), Gaps = 9/141 (6%)
Frame = +2
Query: 194 LHLTQVHFLKLKFLIKILSMSPLLEDLLLKDLQVTDNTLAHDDAAALKPF---PKLLRAD 250
LHL V + +++I++LS P+LE+L + L V + + F PKL+ A+
Sbjct: 134 LHLFGVLLERAEYIIQLLSGCPILEELHAESLIVRNKEWLVSLNFVRQKFPSLPKLITAN 313
Query: 251 VSESSIS-AFLLPLKLFYNVQFLRSQVLLQTLEQDFSTTQFLNLTHMDLIFHDGY----- 304
+++SS S A L L Q LR+++ L +F NLT+M+LI +
Sbjct: 314 ITKSSHSLALFLALLCRAKSQLLRAELDTSHLIH----LKFRNLTNMELILKHNHCEKWN 481
Query: 305 YWISLMKFICACPSLQTLTIH 325
+W L+K + CP LQ LTIH
Sbjct: 482 WW--LLKALKHCPKLQNLTIH 538
>TC82108 similar to GP|21537320|gb|AAM61661.1 unknown {Arabidopsis
thaliana}, partial (7%)
Length = 817
Score = 49.7 bits (117), Expect = 2e-06
Identities = 49/152 (32%), Positives = 70/152 (45%), Gaps = 11/152 (7%)
Frame = +2
Query: 188 LPSLKILHLTQVHFLKLKFLIKILSMSPLLEDLLLKDLQVTDNTLAHDDAAALKPFPKLL 247
LPSLKILHL V F + K+LS P+L++L DL + ++ L+
Sbjct: 20 LPSLKILHLESVCFTYYGHIRKLLSGCPILQELEANDLTIKIPCRVFLGSSQPPSLSNLV 199
Query: 248 RADVSESSISAFLLPLKLFYNVQFLRSQVLLQTLEQDFSTTQFLNLTHMDL---IFHDG- 303
RA++ S+I L N Q +R LL + + F NLTH++L H+
Sbjct: 200 RANI--SNICPMHNLLDRLRNAQHMR---LLAKYSYELHSV-FHNLTHLELRLDFMHNRG 361
Query: 304 -YYWISLMKFICACPSLQTL------TIHNIG 328
W L+K + P LQTL T+HN G
Sbjct: 362 MLKWSWLIKLLQNFPLLQTLIIEEVDTVHNFG 457
>BI265577 similar to GP|10177368|dbj contains similarity to heat shock
transcription factor HSF30~gene_id:K16L22.13, partial
(3%)
Length = 346
Score = 49.7 bits (117), Expect = 2e-06
Identities = 32/86 (37%), Positives = 45/86 (52%), Gaps = 4/86 (4%)
Frame = +3
Query: 28 SKRWRRVWLGMPNA---DRISILPDELLCRILSLLPAKQIMVT-SLLSRRWRSLRPRMTE 83
++RW L A DR+S LPD +LC ILS LP + + T +L+S RWR L +
Sbjct: 12 NRRWLESSLKRRKAEEEDRLSSLPDSILCHILSFLPTRTSVATMTLVSXRWRHLX*HLXV 191
Query: 84 INIDDTSYIHDRDAYDRYYHVIALFL 109
+ DD Y +D YD + A F+
Sbjct: 192 FDFDD--YDYDPHFYDDSFKKFASFV 263
Score = 38.5 bits (88), Expect = 0.004
Identities = 18/35 (51%), Positives = 25/35 (71%), Gaps = 1/35 (2%)
Frame = +3
Query: 1 MSGLPDELLCEILSFLPSEEFPST-SLLSKRWRRV 34
+S LPD +LC ILSFLP+ +T +L+S RWR +
Sbjct: 69 LSSLPDSILCHILSFLPTRTSVATMTLVSXRWRHL 173
>TC79593 similar to GP|10177368|dbj|BAB10659. contains similarity to heat
shock transcription factor HSF30~gene_id:K16L22.13,
partial (6%)
Length = 713
Score = 47.0 bits (110), Expect = 1e-05
Identities = 20/36 (55%), Positives = 29/36 (80%)
Frame = +1
Query: 1 MSGLPDELLCEILSFLPSEEFPSTSLLSKRWRRVWL 36
+S LPD L+ ILSFLPS++ +T++LSKRW+ +WL
Sbjct: 550 ISALPDSLVHHILSFLPSKDATATTVLSKRWKPLWL 657
Score = 45.4 bits (106), Expect = 3e-05
Identities = 23/47 (48%), Positives = 32/47 (67%), Gaps = 1/47 (2%)
Frame = +1
Query: 32 RRVWLGMPNA-DRISILPDELLCRILSLLPAKQIMVTSLLSRRWRSL 77
R++ L +P DRIS LPD L+ ILS LP+K T++LS+RW+ L
Sbjct: 511 RQLQLSIPEEEDRISALPDSLVHHILSFLPSKDATATTVLSKRWKPL 651
>BG581615 similar to GP|10177368|dbj contains similarity to heat shock
transcription factor HSF30~gene_id:K16L22.13, partial
(3%)
Length = 825
Score = 46.2 bits (108), Expect = 2e-05
Identities = 20/36 (55%), Positives = 28/36 (77%), Gaps = 1/36 (2%)
Frame = +3
Query: 1 MSGLPDELLCEILSFLPSEEFPST-SLLSKRWRRVW 35
+SGLPD +LC ILSFLP+ +T +L+S+RWR +W
Sbjct: 321 LSGLPDSILCHILSFLPTRTSVATMNLVSRRWRNLW 428
Score = 45.1 bits (105), Expect = 4e-05
Identities = 25/54 (46%), Positives = 35/54 (64%), Gaps = 3/54 (5%)
Frame = +3
Query: 27 LSKRWRRVWLGMPNA--DRISILPDELLCRILSLLPAKQIMVT-SLLSRRWRSL 77
+SKR ++ L DR+S LPD +LC ILS LP + + T +L+SRRWR+L
Sbjct: 264 ISKRRQKACLAAATGGTDRLSGLPDSILCHILSFLPTRTSVATMNLVSRRWRNL 425
>TC84389 weakly similar to GP|10177368|dbj|BAB10659. contains similarity to
heat shock transcription factor HSF30~gene_id:K16L22.13,
partial (8%)
Length = 692
Score = 45.1 bits (105), Expect = 4e-05
Identities = 20/50 (40%), Positives = 32/50 (64%)
Frame = +3
Query: 42 DRISILPDELLCRILSLLPAKQIMVTSLLSRRWRSLRPRMTEINIDDTSY 91
DRIS LPD ++ ILS +P K +TS+LS+RW L + ++ +D ++
Sbjct: 267 DRISALPDPIIWHILSFVPTKTAAITSILSKRWNPLWLSVLILHFEDETF 416
Score = 43.1 bits (100), Expect = 2e-04
Identities = 18/38 (47%), Positives = 27/38 (70%)
Frame = +3
Query: 1 MSGLPDELLCEILSFLPSEEFPSTSLLSKRWRRVWLGM 38
+S LPD ++ ILSF+P++ TS+LSKRW +WL +
Sbjct: 273 ISALPDPIIWHILSFVPTKTAAITSILSKRWNPLWLSV 386
>AW690594 similar to GP|23498163|emb hypothetical protein {Plasmodium
falciparum 3D7}, partial (10%)
Length = 633
Score = 44.7 bits (104), Expect = 5e-05
Identities = 24/49 (48%), Positives = 31/49 (62%), Gaps = 1/49 (2%)
Frame = +3
Query: 42 DRISILPDELLCRILSLLPAKQIMVT-SLLSRRWRSLRPRMTEINIDDT 89
DRIS LPD LL ILS LP + + T SL SR+WR+L ++ N D+
Sbjct: 255 DRISTLPDPLLLHILSFLPTRTSVATMSLASRKWRNLWEQLQVFNFKDS 401
Score = 38.5 bits (88), Expect = 0.004
Identities = 19/36 (52%), Positives = 25/36 (68%), Gaps = 1/36 (2%)
Frame = +3
Query: 1 MSGLPDELLCEILSFLPSEEFPST-SLLSKRWRRVW 35
+S LPD LL ILSFLP+ +T SL S++WR +W
Sbjct: 261 ISTLPDPLLLHILSFLPTRTSVATMSLASRKWRNLW 368
>AW690384 similar to GP|23496006|gb hypothetical protein {Plasmodium
falciparum 3D7}, partial (1%)
Length = 681
Score = 44.7 bits (104), Expect = 5e-05
Identities = 28/64 (43%), Positives = 34/64 (52%), Gaps = 1/64 (1%)
Frame = +3
Query: 40 NADRISILPDELLCRILSLLPAKQIMVT-SLLSRRWRSLRPRMTEINIDDTSYIHDRDAY 98
N DR+S LPD LLC ILS LP K + T SL+S R+ L ++ D D Y
Sbjct: 270 NKDRLSSLPDSLLCHILSFLPTKTSVCTMSLVSHRYLHL-----------WKHLQDFDFY 416
Query: 99 DRYY 102
D YY
Sbjct: 417 DVYY 428
Score = 38.9 bits (89), Expect = 0.003
Identities = 20/43 (46%), Positives = 28/43 (64%), Gaps = 1/43 (2%)
Frame = +3
Query: 1 MSGLPDELLCEILSFLPSEEFPST-SLLSKRWRRVWLGMPNAD 42
+S LPD LLC ILSFLP++ T SL+S R+ +W + + D
Sbjct: 282 LSSLPDSLLCHILSFLPTKTSVCTMSLVSHRYLHLWKHLQDFD 410
>BI311381 weakly similar to PIR|T46128|T461 hypothetical protein T2J13.140 -
Arabidopsis thaliana, partial (12%)
Length = 803
Score = 44.3 bits (103), Expect = 7e-05
Identities = 19/35 (54%), Positives = 25/35 (71%)
Frame = +1
Query: 1 MSGLPDELLCEILSFLPSEEFPSTSLLSKRWRRVW 35
+S LPD +LC ILSFL + +TS+LSKRW +W
Sbjct: 109 ISTLPDAILCHILSFLETNYADATSVLSKRWNHLW 213
Score = 39.3 bits (90), Expect = 0.002
Identities = 19/36 (52%), Positives = 22/36 (60%)
Frame = +1
Query: 42 DRISILPDELLCRILSLLPAKQIMVTSLLSRRWRSL 77
D IS LPD +LC ILS L TS+LS+RW L
Sbjct: 103 DIISTLPDAILCHILSFLETNYADATSVLSKRWNHL 210
>AI974570 similar to GP|10177368|dbj contains similarity to heat shock
transcription factor HSF30~gene_id:K16L22.13, partial
(3%)
Length = 442
Score = 43.5 bits (101), Expect = 1e-04
Identities = 27/74 (36%), Positives = 38/74 (50%), Gaps = 1/74 (1%)
Frame = +2
Query: 18 SEEFPSTSLLSKRWRRVWLGMPNADRISILPDELLCRILSLLPAKQIMVT-SLLSRRWRS 76
SE + L SKR + DR+S LP+ LLC ILS LP + + T L+SRRW +
Sbjct: 8 SESAMAPELTSKRQKDEANTGGATDRLSSLPEPLLCHILSFLPTRTSIATLPLVSRRWNN 187
Query: 77 LRPRMTEINIDDTS 90
L + + D +
Sbjct: 188LWKHLDAFHFDSNT 229
Score = 39.7 bits (91), Expect = 0.002
Identities = 18/36 (50%), Positives = 25/36 (69%), Gaps = 1/36 (2%)
Frame = +2
Query: 1 MSGLPDELLCEILSFLPSEEFPST-SLLSKRWRRVW 35
+S LP+ LLC ILSFLP+ +T L+S+RW +W
Sbjct: 86 LSSLPEPLLCHILSFLPTRTSIATLPLVSRRWNNLW 193
>BG449234 weakly similar to GP|8979942|gb|A Contains a F-box PF|00646 domain.
{Arabidopsis thaliana}, partial (6%)
Length = 655
Score = 41.6 bits (96), Expect = 4e-04
Identities = 28/91 (30%), Positives = 46/91 (49%)
Frame = +3
Query: 42 DRISILPDELLCRILSLLPAKQIMVTSLLSRRWRSLRPRMTEINIDDTSYIHDRDAYDRY 101
DRIS+L D LL ILS L ++ + T +LS+RW +L + + +D + D + Y+ +
Sbjct: 156 DRISVLADCLLIHILSFLNTREAVQTCILSKRWINLWKTLPTLTLDCYQF-SDCEIYENF 332
Query: 102 YHVIALFLFEMKIHHPIMKTVTILSASPFFN 132
LF+F H + +L + F N
Sbjct: 333 -----LFMFLSLRDHSTALSALLLHNNHFEN 410
Score = 37.4 bits (85), Expect = 0.008
Identities = 19/39 (48%), Positives = 24/39 (60%)
Frame = +3
Query: 1 MSGLPDELLCEILSFLPSEEFPSTSLLSKRWRRVWLGMP 39
+S L D LL ILSFL + E T +LSKRW +W +P
Sbjct: 162 ISVLADCLLIHILSFLNTREAVQTCILSKRWINLWKTLP 278
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.326 0.140 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,858,564
Number of Sequences: 36976
Number of extensions: 209827
Number of successful extensions: 1898
Number of sequences better than 10.0: 118
Number of HSP's better than 10.0 without gapping: 1777
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1882
length of query: 339
length of database: 9,014,727
effective HSP length: 97
effective length of query: 242
effective length of database: 5,428,055
effective search space: 1313589310
effective search space used: 1313589310
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146632.7


