

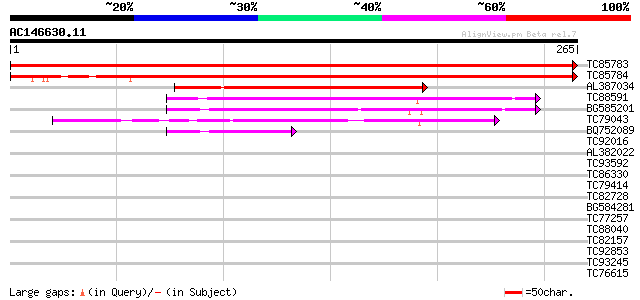
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146630.11 + phase: 0
(265 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85783 homologue to GP|15131688|emb|CAC48323. 2-Cys peroxiredox... 526 e-150
TC85784 similar to GP|15131688|emb|CAC48323. 2-Cys peroxiredoxin... 419 e-118
AL387034 weakly similar to GP|10281259|gb thioredoxin peroxidase... 121 3e-28
TC88591 similar to GP|19423862|gb|AAL88710.1 cys peroxiredoxin {... 91 4e-19
BG585201 weakly similar to GP|14161441|gb thiol-specific antioxi... 80 6e-16
TC79043 homologue to GP|6899842|dbj|BAA90524.1 peroxiredoxin Q {... 74 4e-14
BQ752089 similar to GP|14161441|gb| thiol-specific antioxidant {... 46 1e-05
TC92016 weakly similar to PIR|E70344|E70344 conserved hypothetic... 39 0.002
AL382022 weakly similar to GP|22294918|dbj bacterioferritin comi... 33 0.087
TC93592 weakly similar to GP|13646986|dbj|BAB41080. DNA-binding ... 32 0.25
TC86330 similar to SP|Q43644|NUAM_SOLTU NADH-ubiquinone oxidored... 30 0.96
TC79414 homologue to GP|12597871|gb|AAG60180.1 unknown protein {... 30 0.96
TC82728 similar to GP|7292941|gb|AAF48332.1| CG11071-PA {Drosoph... 30 0.96
BG584281 weakly similar to GP|14209590|dbj hypothetical protein~... 30 1.3
TC77257 weakly similar to GP|14134199|gb|AAK54282.1 fatty acid 9... 29 2.1
TC88040 homologue to SP|Q41266|AOX2_SOYBN Alternative oxidase 2 ... 28 2.8
TC82157 similar to PIR|B32372|B32372 male-specific doublesex pro... 28 2.8
TC92853 similar to GP|20268748|gb|AAM14077.1 unknown protein {Ar... 28 3.6
TC93245 similar to PIR|T05995|T05995 hypothetical protein F17M5.... 28 3.6
TC76615 homologue to SP|O22582|H2B_GOSHI Histone H2B. [Upland co... 28 4.8
>TC85783 homologue to GP|15131688|emb|CAC48323. 2-Cys peroxiredoxin {Pisum
sativum}, partial (91%)
Length = 1136
Score = 526 bits (1355), Expect = e-150
Identities = 265/265 (100%), Positives = 265/265 (100%)
Frame = +3
Query: 1 MACCSAPSASLLSSNPNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFTSSR 60
MACCSAPSASLLSSNPNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFTSSR
Sbjct: 48 MACCSAPSASLLSSNPNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFTSSR 227
Query: 61 RSFVVRASSELPLVGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPT 120
RSFVVRASSELPLVGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPT
Sbjct: 228 RSFVVRASSELPLVGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPT 407
Query: 121 EITAFSDRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISK 180
EITAFSDRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISK
Sbjct: 408 EITAFSDRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISK 587
Query: 181 SYGVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDEVC 240
SYGVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDEVC
Sbjct: 588 SYGVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDEVC 767
Query: 241 PAGWKPGEKSMKPDPKLSKEYFSAV 265
PAGWKPGEKSMKPDPKLSKEYFSAV
Sbjct: 768 PAGWKPGEKSMKPDPKLSKEYFSAV 842
>TC85784 similar to GP|15131688|emb|CAC48323. 2-Cys peroxiredoxin {Pisum
sativum}, partial (86%)
Length = 1114
Score = 419 bits (1076), Expect = e-118
Identities = 225/273 (82%), Positives = 239/273 (87%), Gaps = 8/273 (2%)
Frame = +2
Query: 1 MACCSAPSA--SLLSS-NP-NILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNR- 55
MAC + ++ SL SS NP + LFSPKL P S+LSIPN SLPK + LS R
Sbjct: 134 MACSATTTSASSLFSSLNPKSSLFSPKL---PSSSTLSIPN---SLPKPFSLPSLSFTRP 295
Query: 56 ---FTSSRRSFVVRASSELPLVGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPL 112
+S R SF+V+ASSELPLVGNAAPDFEAEAVFDQEFI VKLS+YIGKKYVILFFYPL
Sbjct: 296 SLHHSSRRSSFLVKASSELPLVGNAAPDFEAEAVFDQEFINVKLSDYIGKKYVILFFYPL 475
Query: 113 DFTFVCPTEITAFSDRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVS 172
DFTFVCPTEITAFSDRHAEF +NTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVS
Sbjct: 476 DFTFVCPTEITAFSDRHAEFEAINTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVS 655
Query: 173 DVTKSISKSYGVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYV 232
DVTKSISKSYGVLIPDQGIALRGLFIIDKEG+IQHSTINNLGIGRSVDETKRTLQALQYV
Sbjct: 656 DVTKSISKSYGVLIPDQGIALRGLFIIDKEGIIQHSTINNLGIGRSVDETKRTLQALQYV 835
Query: 233 QENPDEVCPAGWKPGEKSMKPDPKLSKEYFSAV 265
QENPDEVCPAGWKPGEKSMKPDPKLSK+YF+AV
Sbjct: 836 QENPDEVCPAGWKPGEKSMKPDPKLSKDYFAAV 934
>AL387034 weakly similar to GP|10281259|gb thioredoxin peroxidase 3
{Schistosoma mansoni}, partial (54%)
Length = 376
Score = 121 bits (303), Expect = 3e-28
Identities = 55/118 (46%), Positives = 81/118 (68%)
Frame = +2
Query: 78 APDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPTEITAFSDRHAEFAELNT 137
AP +EA+AV D + + L ++ KY+++ FYPLDFTFVCPTE+ AFSDR EF ++NT
Sbjct: 23 APHWEAKAVVDGDVKTLSLKDF-ASKYLVMVFYPLDFTFVCPTELIAFSDRVQEFRDINT 199
Query: 138 EILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISKSYGVLIPDQGIALRG 195
E++G+S DS +H+AW + R GGLG + PL++D+ K I++ YG + + G RG
Sbjct: 200 EVVGISCDSHHAHIAWDKIPRNQGGLGGIKIPLIADLKKEITRRYGCMFEETGHPFRG 373
>TC88591 similar to GP|19423862|gb|AAL88710.1 cys peroxiredoxin {Xerophyta
viscosa}, partial (92%)
Length = 815
Score = 91.3 bits (225), Expect = 4e-19
Identities = 55/180 (30%), Positives = 89/180 (48%), Gaps = 5/180 (2%)
Frame = +2
Query: 74 VGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPTEITAFSDRHAEFA 133
+G+ PD E + K+KL + + ILF +P DFT VC TE+ + +EF
Sbjct: 35 IGDTIPDLEVDTTQG----KIKLHHFCSDSWTILFSHPGDFTPVCTTELGKMAQYASEFN 202
Query: 134 ELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISKSYGVLIPDQ---- 189
+ +LG+S D + SH W++ +NYP++SD + I K ++ PD+
Sbjct: 203 KRGVMLLGMSCDDLESHKEWIKDIEAHTPGAKVNYPIISDPKREIIKQLNMVDPDEKDSN 382
Query: 190 -GIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDEVCPAGWKPGE 248
+ R L I+ + I+ S + GR++DE R +++LQ + PA WKPGE
Sbjct: 383 GNLPSRALHIVGPDKKIKLSFLYPAQTGRNMDEVLRVVESLQKASKY-KIATPANWKPGE 559
>BG585201 weakly similar to GP|14161441|gb thiol-specific antioxidant
{Ajellomyces capsulatus}, partial (92%)
Length = 731
Score = 80.5 bits (197), Expect = 6e-16
Identities = 49/184 (26%), Positives = 94/184 (50%), Gaps = 9/184 (4%)
Frame = +1
Query: 74 VGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPTEITAFSDRHAEFA 133
+G+ AP+F+A+ + + E+I + +LF +P DFT VC TE+ + RH +F
Sbjct: 10 LGDEAPNFKAKTTKGE----IDFHEFIKNSWAVLFSHPADFTPVCTTELGEVARRHKDFT 177
Query: 134 ELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISKSYGVL-------I 186
+++G+S + + H WV+ + + +++P+++D +++S Y +L +
Sbjct: 178 TRGVKVIGLSANELKDHEKWVEDINEVNSV-TVDFPIIADSDRTVSTLYDMLDHLDATNV 354
Query: 187 PDQGI--ALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDEVCPAGW 244
+GI +R +FIID + I+ GR+ DE R + +LQ + + PA W
Sbjct: 355 DTKGIPFTVRNVFIIDPKKKIRLMITYPASTGRNFDEVIRVIDSLQ-LGDKHRITTPANW 531
Query: 245 KPGE 248
G+
Sbjct: 532 NKGD 543
>TC79043 homologue to GP|6899842|dbj|BAA90524.1 peroxiredoxin Q {Sedum
lineare}, partial (81%)
Length = 935
Score = 74.3 bits (181), Expect = 4e-14
Identities = 65/210 (30%), Positives = 100/210 (46%), Gaps = 1/210 (0%)
Frame = +3
Query: 21 SPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFTSSRRSFVVRASSELPLVGNAAPD 80
S L+S P S S+S TS P S S +R F+ S+ G+ P
Sbjct: 120 STSLTSAPFNSQFFGLKLSHSSISKTTSSPPS-----SFKRGFIFAKVSK----GSKPPA 272
Query: 81 FEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPTEITAFSDRHAEFAELNTEIL 140
F + DQ+ V LS+Y GK V+++FYP D T C + AF D + +F + E++
Sbjct: 273 FTLK---DQDGKTVSLSKYKGKP-VVVYFYPADETPGCTKQACAFRDSYEKFKKAGAEVV 440
Query: 141 GVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISKSYGVLIPDQG-IALRGLFII 199
G+S D SH A+ + + L + L+SD + K +GV G + R +++
Sbjct: 441 GISGDDSSSHKAFAKKYK-------LPFTLLSDEGNKVRKEWGVPGDFFGSLPGRETYVL 599
Query: 200 DKEGVIQHSTINNLGIGRSVDETKRTLQAL 229
DK GV+Q N + +DET + LQ+L
Sbjct: 600 DKNGVVQLVYNNQFQPEKHIDETLKLLQSL 689
>BQ752089 similar to GP|14161441|gb| thiol-specific antioxidant {Ajellomyces
capsulatus}, partial (30%)
Length = 204
Score = 46.2 bits (108), Expect = 1e-05
Identities = 23/61 (37%), Positives = 36/61 (58%)
Frame = +3
Query: 74 VGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPTEITAFSDRHAEFA 133
+G AP+F+AE + E+IG +V+LF +P D+T VC TE+ AF+ EF+
Sbjct: 33 LGTVAPNFKAETTNGP----IDFHEFIGDSWVVLFSHPEDYTPVCTTELGAFAKLEPEFS 200
Query: 134 E 134
+
Sbjct: 201 K 203
>TC92016 weakly similar to PIR|E70344|E70344 conserved hypothetical protein
aq_495 - Aquifex aeolicus, partial (16%)
Length = 629
Score = 38.9 bits (89), Expect = 0.002
Identities = 20/56 (35%), Positives = 31/56 (54%), Gaps = 1/56 (1%)
Frame = +1
Query: 88 DQEFIKVKLSEYIGK-KYVILFFYPLDFTFVCPTEITAFSDRHAEFAELNTEILGV 142
+Q+ V L +G K I+FFYP D T+ C E+ F D ++ F+E +LG+
Sbjct: 91 NQDNEDVNLDTMVGTGKPSIVFFYPKDETYGCTQEVCHFRDSYSVFSEAGATVLGI 258
>AL382022 weakly similar to GP|22294918|dbj bacterioferritin comigratory
protein {Thermosynechococcus elongatus BP-1}, partial
(16%)
Length = 392
Score = 33.5 bits (75), Expect = 0.087
Identities = 23/67 (34%), Positives = 32/67 (47%)
Frame = +3
Query: 140 LGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISKSYGVLIPDQGIALRGLFII 199
LG+S D V H + T R L +PL+SD K Y V + R F+I
Sbjct: 24 LGISSDPVDKHKRFSDTQR-------LQFPLLSDPNGEARKLYEVPKTLGFLPGRASFLI 182
Query: 200 DKEGVIQ 206
DK+G+I+
Sbjct: 183 DKDGIIR 203
>TC93592 weakly similar to GP|13646986|dbj|BAB41080. DNA-binding protein DF1
{Pisum sativum}, partial (11%)
Length = 622
Score = 32.0 bits (71), Expect = 0.25
Identities = 19/54 (35%), Positives = 29/54 (53%)
Frame = -1
Query: 6 APSASLLSSNPNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFTSS 59
+P AS+L + + F S PP L + ++ S+SLP L +LP N F+ S
Sbjct: 478 SPRASMLLLSLSFPFPSSQSFPPFLYASALAFPSSSLPNLLQNLPAFSNSFSFS 317
>TC86330 similar to SP|Q43644|NUAM_SOLTU NADH-ubiquinone oxidoreductase 75
kDa subunit mitochondrial precursor (EC 1.6.5.3) (EC
1.6.99.3), partial (95%)
Length = 2619
Score = 30.0 bits (66), Expect = 0.96
Identities = 30/106 (28%), Positives = 52/106 (48%)
Frame = +3
Query: 13 SSNPNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFTSSRRSFVVRASSELP 72
S+NPN +P +P LSS+S+P + + + +P++L RF +S +P
Sbjct: 177 SANPNPPLNP---NPNSLSSISLPVHPSLALESISPIPMTLLRF----------SSMVIP 317
Query: 73 LVGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVC 118
L F+ +++F F +V+L I +VI+ +PL VC
Sbjct: 318 L------KFQKDSLF---FKRVRLLVLIFLDFVIIADFPLLEIVVC 428
>TC79414 homologue to GP|12597871|gb|AAG60180.1 unknown protein {Arabidopsis
thaliana}, partial (36%)
Length = 1700
Score = 30.0 bits (66), Expect = 0.96
Identities = 22/60 (36%), Positives = 33/60 (54%), Gaps = 6/60 (10%)
Frame = +2
Query: 16 PNILFSPK--LSSPPHLSSLSIPNASNSLPKLRTSLPLS----LNRFTSSRRSFVVRASS 69
P +FS K LS+PP ++LS P A+++ PKL+ S N S SFV+ ++S
Sbjct: 683 PLSIFSSK*LLSTPPSPTTLSTPPATSTTPKLKPRTWFS*TY*RNSCLSKPSSFVIPSTS 862
>TC82728 similar to GP|7292941|gb|AAF48332.1| CG11071-PA {Drosophila
melanogaster}, partial (2%)
Length = 734
Score = 30.0 bits (66), Expect = 0.96
Identities = 21/59 (35%), Positives = 29/59 (48%), Gaps = 10/59 (16%)
Frame = -1
Query: 5 SAPSASLLSSNPNILFSPKLSSPPHL----------SSLSIPNASNSLPKLRTSLPLSL 53
+A S+++ SS+PN L SPP L SSL IP+ L K ++PL L
Sbjct: 500 TASSSTITSSSPNF*SKITLPSPPSLSSSSHQRENESSLRIPDPELPLSKSSIAIPLKL 324
>BG584281 weakly similar to GP|14209590|dbj hypothetical protein~similar to
Arabidopsis thaliana chromosome 3 MMM17.14, partial
(28%)
Length = 761
Score = 29.6 bits (65), Expect = 1.3
Identities = 19/48 (39%), Positives = 24/48 (49%), Gaps = 2/48 (4%)
Frame = +2
Query: 10 SLLSSNPNILFSPKLSSPPHLSSLSIPNA--SNSLPKLRTSLPLSLNR 55
SLL I+ + SSPPH SSL N+ N + + T PLS R
Sbjct: 23 SLLCYESFIIMNSSSSSPPHYSSLPSSNSIPPNFISRATTPKPLSATR 166
>TC77257 weakly similar to GP|14134199|gb|AAK54282.1 fatty acid
9-hydroperoxide lyase {Cucumis melo}, partial (81%)
Length = 1989
Score = 28.9 bits (63), Expect = 2.1
Identities = 21/82 (25%), Positives = 40/82 (48%)
Frame = +3
Query: 1 MACCSAPSASLLSSNPNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFTSSR 60
+A +A S++ N + F + ++S + N+ ++ KL ++LPL TSS
Sbjct: 477 IASSNASSSTSFHQNTTLSFHSFKLT*LNISPILKKNSLENIKKLASTLPLEA*PSTSSS 656
Query: 61 RSFVVRASSELPLVGNAAPDFE 82
S + + ++L LV P F+
Sbjct: 657 NSSLTKTLAKLKLVTLVQP*FK 722
>TC88040 homologue to SP|Q41266|AOX2_SOYBN Alternative oxidase 2
mitochondrial precursor (EC 1.-.-.-). [Soybean] {Glycine
max}, partial (82%)
Length = 1678
Score = 28.5 bits (62), Expect = 2.8
Identities = 19/55 (34%), Positives = 29/55 (52%), Gaps = 4/55 (7%)
Frame = -3
Query: 10 SLLSSNPNILFSPKLSS----PPHLSSLSIPNASNSLPKLRTSLPLSLNRFTSSR 60
SL S +PN P+ SS PP S S + SLP + +++PL+ +SS+
Sbjct: 707 SLASKSPNNSTQPRYSSFRNSPPSSSRSSRSISPPSLPPMASTVPLAKTSSSSSK 543
>TC82157 similar to PIR|B32372|B32372 male-specific doublesex protein -
fruit fly (Drosophila melanogaster), partial (3%)
Length = 921
Score = 28.5 bits (62), Expect = 2.8
Identities = 19/56 (33%), Positives = 30/56 (52%)
Frame = +3
Query: 13 SSNPNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFTSSRRSFVVRAS 68
S+N N+ + +P + SS S+ N SNSLPK +L + +SS F + A+
Sbjct: 363 SNNSNVPLKNAIPTPKN-SSFSMSNMSNSLPKPVNTLQSGSSLGSSSGSGFNLNAN 527
>TC92853 similar to GP|20268748|gb|AAM14077.1 unknown protein {Arabidopsis
thaliana}, partial (21%)
Length = 761
Score = 28.1 bits (61), Expect = 3.6
Identities = 20/47 (42%), Positives = 27/47 (56%)
Frame = -2
Query: 5 SAPSASLLSSNPNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPL 51
S PS+SL+SS+ + SS SS S+P++S SL SLPL
Sbjct: 451 SLPSSSLVSSSSS-------SSSSSSSSFSLPSSSLSLSLPLVSLPL 332
>TC93245 similar to PIR|T05995|T05995 hypothetical protein F17M5.200 -
Arabidopsis thaliana, partial (21%)
Length = 778
Score = 28.1 bits (61), Expect = 3.6
Identities = 23/64 (35%), Positives = 31/64 (47%), Gaps = 3/64 (4%)
Frame = +2
Query: 14 SNPNILFSPKLSSPPHLSSL---SIPNASNSLPKLRTSLPLSLNRFTSSRRSFVVRASSE 70
S+P+ LF+ SPP LSSL S PN N + P+S F + S S+E
Sbjct: 206 SSPSSLFN----SPPALSSLSPSSSPNNHNPPSSIPPPAPVSSTTFLPEKLSSRSLISAE 373
Query: 71 LPLV 74
L +V
Sbjct: 374 LAMV 385
>TC76615 homologue to SP|O22582|H2B_GOSHI Histone H2B. [Upland cotton]
{Gossypium hirsutum}, complete
Length = 685
Score = 27.7 bits (60), Expect = 4.8
Identities = 22/73 (30%), Positives = 33/73 (45%), Gaps = 1/73 (1%)
Frame = -2
Query: 3 CCSAPSASLLSSNPNILFSPKLSSPPHLSSLS-IPNASNSLPKLRTSLPLSLNRFTSSRR 61
C P++ L + S L L+ +P ++SLP + L LS R+TSS +
Sbjct: 297 CLGEPASEL*RCRSCMFLHSSCFSSSCLRQLNHLPLVASSLPLASSLLELSRQRWTSSPQ 118
Query: 62 SFVVRASSELPLV 74
+S LPLV
Sbjct: 117 LVSSPLASSLPLV 79
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.134 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,170,693
Number of Sequences: 36976
Number of extensions: 112576
Number of successful extensions: 949
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 938
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 946
length of query: 265
length of database: 9,014,727
effective HSP length: 94
effective length of query: 171
effective length of database: 5,538,983
effective search space: 947166093
effective search space used: 947166093
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146630.11


