

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146590.2 + phase: 0
(313 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
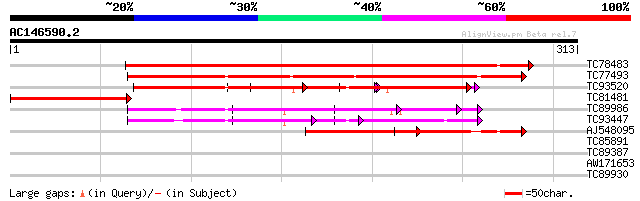
Score E
Sequences producing significant alignments: (bits) Value
TC78483 weakly similar to SP|P08688|ALB2_PEA Albumin 2 (PA2). [G... 243 7e-65
TC77493 weakly similar to GP|20159765|gb|AAM12036.1 anther-speci... 196 7e-51
TC93520 weakly similar to GP|20159765|gb|AAM12036.1 anther-speci... 102 4e-42
TC81481 PIR|B44916|S27514 mosquitocidal toxin 21 - Bacillus spha... 141 4e-34
TC89986 weakly similar to SP|P08688|ALB2_PEA Albumin 2 (PA2). [G... 107 8e-24
TC93447 79 3e-15
AJ548095 58 4e-09
TC85891 similar to PIR|A86386|A86386 probable DNA-binding protei... 28 3.5
TC89387 similar to GP|17979249|gb|AAL49941.1 AT3g04610/F7O18_9 {... 28 4.6
AW171653 28 6.0
TC89930 similar to PIR|T47694|T47694 probable serine/threonine-s... 27 7.8
>TC78483 weakly similar to SP|P08688|ALB2_PEA Albumin 2 (PA2). [Garden pea]
{Pisum sativum}, partial (96%)
Length = 997
Score = 243 bits (620), Expect = 7e-65
Identities = 116/226 (51%), Positives = 158/226 (69%), Gaps = 1/226 (0%)
Frame = +2
Query: 65 YINAAFRASANNEAYLFMNNEYVLINYARGSTNDYIINGPLYICDGYPSLARTPFGEHGI 124
YINA FR+S NEAY F+N++ ++++YA G++ND I++GP+++ G+PSL T FG +GI
Sbjct: 77 YINAVFRSSRKNEAYFFINDKLLVLDYAPGTSNDKILHGPVFVRHGFPSLDNTKFGSNGI 256
Query: 125 DCAFDTDKTQAYIFSANLCALIDYAPGTTNDKILSGPMKITDMFPFFKDTVFERGLDAAF 184
DCAFDTD +A++F LCA I+Y P T DKI+SGPMKI +MFPF + T FE G+DAAF
Sbjct: 257 DCAFDTDDNEAFVFYQGLCAKIEYVPQTDKDKIISGPMKIAEMFPFLEGTGFEHGIDAAF 436
Query: 185 RAHSSNEAYLFRGGHYALINYSSKRLIH-IETIRHGFPSLIGTVFENGLEAAFASHGTKE 243
R+ + E YLFRG YA I+Y + L+ I I GF T+FE G++AAFASH E
Sbjct: 437 RSTLNKEVYLFRGDKYARIDYGTNSLVQIIRDINSGFTCFRDTIFEKGIDAAFASHIPNE 616
Query: 244 AYLFKGEYYANIYYAPGSTDDYLIGGRVKLILSNWPSLKKILPRNN 289
AYLFKG+ + I + PG +DDY++GG V+ L W SL+ I+P N
Sbjct: 617 AYLFKGDNFVRITFTPGRSDDYIMGG-VRSTLDVWKSLRDIIPLKN 751
>TC77493 weakly similar to GP|20159765|gb|AAM12036.1 anther-specific protein
{Pisum sativum}, partial (25%)
Length = 1177
Score = 196 bits (499), Expect = 7e-51
Identities = 100/220 (45%), Positives = 143/220 (64%)
Frame = +1
Query: 66 INAAFRASANNEAYLFMNNEYVLINYARGSTNDYIINGPLYICDGYPSLARTPFGEHGID 125
I+ AFR+S NE Y+F + ++NYA G+ D ++ P++ DG+P L TPF E +D
Sbjct: 91 IDTAFRSSKLNECYVFKKKRFTVVNYAPGAKKDRVVKRPIHTSDGFPFLDETPF-EKVVD 267
Query: 126 CAFDTDKTQAYIFSANLCALIDYAPGTTNDKILSGPMKITDMFPFFKDTVFERGLDAAFR 185
CAF+T+ T+A IFS N CA ID+AP + + ++ GP+ IT MF K T E G+DAA +
Sbjct: 268 CAFETEGTEAIIFSGNKCAKIDFAPDSDTE-LVDGPVPITTMFHCLKGTFLENGIDAAIK 444
Query: 186 AHSSNEAYLFRGGHYALINYSSKRLIHIETIRHGFPSLIGTVFENGLEAAFASHGTKEAY 245
N +LF+G Y ++Y SK ++ +IR GF SL+GTVFENG++AAFASH +AY
Sbjct: 445 WTGHN-VFLFKGNEYVRMDYHSKTIVTHYSIRSGFKSLLGTVFENGIDAAFASHVPNQAY 621
Query: 246 LFKGEYYANIYYAPGSTDDYLIGGRVKLILSNWPSLKKIL 285
+FKGEYYA I + + D ++ GRVKL WP+L+ +L
Sbjct: 622 VFKGEYYARIDIS--NEGDCIVNGRVKLFHDEWPALQGLL 735
>TC93520 weakly similar to GP|20159765|gb|AAM12036.1 anther-specific protein
{Pisum sativum}, partial (26%)
Length = 659
Score = 102 bits (254), Expect(2) = 4e-42
Identities = 51/101 (50%), Positives = 70/101 (68%), Gaps = 5/101 (4%)
Frame = +3
Query: 69 AFRASANNEAYLFMNNEYVLINYARGSTNDYIINGPLYICDGYPSLARTPFGEHGIDCAF 128
AFR+ +NE Y+F++++YV++NYA G+ I+ GPL+I G+ LARTPF EHGIDCAF
Sbjct: 63 AFRSFKHNECYVFVDDKYVVLNYAPGAKKHEILKGPLHIVAGFAMLARTPF-EHGIDCAF 239
Query: 129 DTDKTQAYIFSANLCALIDYAPGTTND-----KILSGPMKI 164
+T +A+IFS N CALI Y P + +IL+GP KI
Sbjct: 240 ETQHNEAFIFSGNHCALITYGPHVPHHQHHPARILAGPKKI 362
Score = 86.7 bits (213), Expect(2) = 4e-42
Identities = 45/93 (48%), Positives = 58/93 (61%)
Frame = +1
Query: 163 KITDMFPFFKDTVFERGLDAAFRAHSSNEAYLFRGGHYALINYSSKRLIHIETIRHGFPS 222
K+ MF T FE G+DAA R LF+G YAL++Y S R++ IR GF +
Sbjct: 358 KLQHMFTCLHGTAFEHGIDAAIRT-LDKRVLLFKGNAYALMDYHSNRVLANHYIRTGFKT 534
Query: 223 LIGTVFENGLEAAFASHGTKEAYLFKGEYYANI 255
L+GTVFENG++AAF S EAY+FK +YYA I
Sbjct: 535 LVGTVFENGIDAAFKSDKKDEAYIFKNQYYACI 633
Score = 59.3 bits (142), Expect = 2e-09
Identities = 29/72 (40%), Positives = 40/72 (55%)
Frame = +3
Query: 134 QAYIFSANLCALIDYAPGTTNDKILSGPMKITDMFPFFKDTVFERGLDAAFRAHSSNEAY 193
+ Y+F + +++YAPG +IL GP+ I F T FE G+D AF NEA+
Sbjct: 87 ECYVFVDDKYVVLNYAPGAKKHEILKGPLHIVAGFAMLARTPFEHGIDCAFET-QHNEAF 263
Query: 194 LFRGGHYALINY 205
+F G H ALI Y
Sbjct: 264 IFSGNHCALITY 299
Score = 58.2 bits (139), Expect = 4e-09
Identities = 33/84 (39%), Positives = 48/84 (56%)
Frame = +1
Query: 121 EHGIDCAFDTDKTQAYIFSANLCALIDYAPGTTNDKILSGPMKITDMFPFFKDTVFERGL 180
EHGID A T + +F N AL+DY ++++L+ I F TVFE G+
Sbjct: 400 EHGIDAAIRTLDKRVLLFKGNAYALMDYH----SNRVLANHY-IRTGFKTLVGTVFENGI 564
Query: 181 DAAFRAHSSNEAYLFRGGHYALIN 204
DAAF++ +EAY+F+ +YA IN
Sbjct: 565 DAAFKSDKKDEAYIFKNQYYACIN 636
Score = 47.0 bits (110), Expect = 9e-06
Identities = 30/85 (35%), Positives = 43/85 (50%), Gaps = 8/85 (9%)
Frame = +3
Query: 183 AFRAHSSNEAYLFRGGHYALINYSS--------KRLIHIETIRHGFPSLIGTVFENGLEA 234
AFR+ NE Y+F Y ++NY+ K +HI GF L T FE+G++
Sbjct: 63 AFRSFKHNECYVFVDDKYVVLNYAPGAKKHEILKGPLHIVA---GFAMLARTPFEHGIDC 233
Query: 235 AFASHGTKEAYLFKGEYYANIYYAP 259
AF + EA++F G + A I Y P
Sbjct: 234 AFETQ-HNEAFIFSGNHCALITYGP 305
Score = 31.2 bits (69), Expect = 0.54
Identities = 13/29 (44%), Positives = 19/29 (64%)
Frame = +1
Query: 66 INAAFRASANNEAYLFMNNEYVLINYARG 94
I+AAF++ +EAY+F N Y IN +G
Sbjct: 562 IDAAFKSDKKDEAYIFKNQYYACINIDQG 648
Score = 27.3 bits (59), Expect = 7.8
Identities = 12/40 (30%), Positives = 21/40 (52%)
Frame = +3
Query: 235 AFASHGTKEAYLFKGEYYANIYYAPGSTDDYLIGGRVKLI 274
AF S E Y+F + Y + YAPG+ ++ G + ++
Sbjct: 63 AFRSFKHNECYVFVDDKYVVLNYAPGAKKHEILKGPLHIV 182
>TC81481 PIR|B44916|S27514 mosquitocidal toxin 21 - Bacillus sphaericus,
partial (1%)
Length = 880
Score = 141 bits (355), Expect = 4e-34
Identities = 67/67 (100%), Positives = 67/67 (100%)
Frame = +2
Query: 1 MDLQFTIHTPITFYIDDNGIRRELPEETYTPHERDQVQKRDVSSSQDNEEVELHSYGAVE 60
MDLQFTIHTPITFYIDDNGIRRELPEETYTPHERDQVQKRDVSSSQDNEEVELHSYGAVE
Sbjct: 680 MDLQFTIHTPITFYIDDNGIRRELPEETYTPHERDQVQKRDVSSSQDNEEVELHSYGAVE 859
Query: 61 NIPIYIN 67
NIPIYIN
Sbjct: 860 NIPIYIN 880
>TC89986 weakly similar to SP|P08688|ALB2_PEA Albumin 2 (PA2). [Garden pea]
{Pisum sativum}, partial (27%)
Length = 560
Score = 107 bits (266), Expect = 8e-24
Identities = 58/153 (37%), Positives = 89/153 (57%), Gaps = 2/153 (1%)
Frame = +2
Query: 66 INAAFRASANNEAYLFMNNEYVLINYARGSTNDYIINGPLYICDGYPSLARTPFGEHGID 125
I AAFR+S NE YLF+ YV+++Y + +I+GP+ I G+P ++T F + GID
Sbjct: 86 IGAAFRSSKPNECYLFVKERYVILDYY--TLRKDLISGPVEIDAGFPMFSKTIF-KRGID 256
Query: 126 CAFDTDKTQAYIFSANLCALIDYAP--GTTNDKILSGPMKITDMFPFFKDTVFERGLDAA 183
+F+T+ AY FS + C DYAP G +I+ GP++I +MFP T F G+DAA
Sbjct: 257 SSFETEGNVAYFFSKSQCVKTDYAPRSGPAAARIVIGPIEIVEMFPSLVTTPFANGIDAA 436
Query: 184 FRAHSSNEAYLFRGGHYALINYSSKRLIHIETI 216
R S +A LF+G ++++ + +E I
Sbjct: 437 LRFPGSKDAVLFKGNMCGILDFKYNHVYEVENI 535
Score = 71.2 bits (173), Expect = 5e-13
Identities = 46/134 (34%), Positives = 69/134 (51%), Gaps = 8/134 (5%)
Frame = +2
Query: 124 IDCAFDTDK-TQAYIFSANLCALIDYAPGTTNDKILSGPMKITDMFPFFKDTVFERGLDA 182
I AF + K + Y+F ++DY T ++SGP++I FP F T+F+RG+D+
Sbjct: 86 IGAAFRSSKPNECYLFVKERYVILDYY--TLRKDLISGPVEIDAGFPMFSKTIFKRGIDS 259
Query: 183 AFRAHSSNEAYLFRGGHYALINYSSKR-------LIHIETIRHGFPSLIGTVFENGLEAA 235
+F N AY F +Y+ + +I I FPSL+ T F NG++AA
Sbjct: 260 SFETEG-NVAYFFSKSQCVKTDYAPRSGPAAARIVIGPIEIVEMFPSLVTTPFANGIDAA 436
Query: 236 FASHGTKEAYLFKG 249
G+K+A LFKG
Sbjct: 437 LRFPGSKDAVLFKG 478
Score = 46.6 bits (109), Expect = 1e-05
Identities = 29/85 (34%), Positives = 42/85 (49%), Gaps = 3/85 (3%)
Frame = +2
Query: 180 LDAAFRAHSSNEAYLFRGGHYALINYSSKRLIHIE---TIRHGFPSLIGTVFENGLEAAF 236
+ AAFR+ NE YLF Y +++Y + R I I GFP T+F+ G++++F
Sbjct: 86 IGAAFRSSKPNECYLFVKERYVILDYYTLRKDLISGPVEIDAGFPMFSKTIFKRGIDSSF 265
Query: 237 ASHGTKEAYLFKGEYYANIYYAPGS 261
+ G AY F YAP S
Sbjct: 266 ETEG-NVAYFFSKSQCVKTDYAPRS 337
>TC93447
Length = 343
Score = 78.6 bits (192), Expect = 3e-15
Identities = 43/106 (40%), Positives = 64/106 (59%), Gaps = 2/106 (1%)
Frame = +2
Query: 66 INAAFRASANNEAYLFMNNEYVLINYARGSTNDYIINGPLYICDGYPSLARTPFGEHGID 125
I AAFR+S E YLF+ YV++NY N +I+GP+ I G+P ++T F + G+D
Sbjct: 38 IGAAFRSSRPYECYLFVKERYVILNY----HNKELISGPVDIEAGFPMFSKTIF-KRGVD 202
Query: 126 CAFDTDKTQAYIFSANLCALIDYAP--GTTNDKILSGPMKITDMFP 169
+F+T+ AY F + C DYAP G +I+ GP++I +MFP
Sbjct: 203 SSFETEGNVAYFFYKSKCVKTDYAPRSGPAAARIVIGPVEIVEMFP 340
Score = 48.9 bits (115), Expect = 2e-06
Identities = 30/83 (36%), Positives = 42/83 (50%), Gaps = 1/83 (1%)
Frame = +2
Query: 180 LDAAFRAHSSNEAYLFRGGHYALINYSSKRLIHIET-IRHGFPSLIGTVFENGLEAAFAS 238
+ AAFR+ E YLF Y ++NY +K LI I GFP T+F+ G++++F +
Sbjct: 38 IGAAFRSSRPYECYLFVKERYVILNYHNKELISGPVDIEAGFPMFSKTIFKRGVDSSFET 217
Query: 239 HGTKEAYLFKGEYYANIYYAPGS 261
G AY F YAP S
Sbjct: 218 EG-NVAYFFYKSKCVKTDYAPRS 283
Score = 42.7 bits (99), Expect = 2e-04
Identities = 24/73 (32%), Positives = 40/73 (53%), Gaps = 1/73 (1%)
Frame = +2
Query: 124 IDCAFDTDKT-QAYIFSANLCALIDYAPGTTNDKILSGPMKITDMFPFFKDTVFERGLDA 182
I AF + + + Y+F +++Y N +++SGP+ I FP F T+F+RG+D+
Sbjct: 38 IGAAFRSSRPYECYLFVKERYVILNYH----NKELISGPVDIEAGFPMFSKTIFKRGVDS 205
Query: 183 AFRAHSSNEAYLF 195
+F N AY F
Sbjct: 206 SFET-EGNVAYFF 241
>AJ548095
Length = 593
Score = 58.2 bits (139), Expect = 4e-09
Identities = 28/73 (38%), Positives = 47/73 (64%)
Frame = -2
Query: 213 IETIRHGFPSLIGTVFENGLEAAFASHGTKEAYLFKGEYYANIYYAPGSTDDYLIGGRVK 272
+E I + +P+ + TVFE G++AAF +HG E ++FKGE+ A + + IGG +K
Sbjct: 571 VENITYHYPTFVDTVFEEGIDAAFCAHGGNEIFIFKGEHCARV-----NLFGQFIGG-IK 410
Query: 273 LILSNWPSLKKIL 285
I ++WP+L+ I+
Sbjct: 409 RIDADWPTLRGII 371
Score = 56.6 bits (135), Expect = 1e-08
Identities = 28/64 (43%), Positives = 40/64 (61%)
Frame = -2
Query: 164 ITDMFPFFKDTVFERGLDAAFRAHSSNEAYLFRGGHYALINYSSKRLIHIETIRHGFPSL 223
IT +P F DTVFE G+DAAF AH NE ++F+G H A +N + + I+ I +P+L
Sbjct: 562 ITYHYPTFVDTVFEEGIDAAFCAHGGNEIFIFKGEHCARVNLFGQFIGGIKRIDADWPTL 383
Query: 224 IGTV 227
G +
Sbjct: 382 RGII 371
>TC85891 similar to PIR|A86386|A86386 probable DNA-binding protein RAV2
[imported] - Arabidopsis thaliana, partial (57%)
Length = 2083
Score = 28.5 bits (62), Expect = 3.5
Identities = 22/78 (28%), Positives = 31/78 (39%), Gaps = 1/78 (1%)
Frame = -2
Query: 171 FKDTVFERGLDAAFRAHSSNEAYLFRGGHYALINYSSKRLIHIETIRHGFPSLIG-TVFE 229
FK F +H++ A F G ++ + T R FP+ +G TVF
Sbjct: 1323 FKSKPFAAAAAVETPSHATATATAFNGKCFSACCFGI-------TNRFNFPTSLGVTVFS 1165
Query: 230 NGLEAAFASHGTKEAYLF 247
N A ASH +E F
Sbjct: 1164 NNCSRALASHAPRETAPF 1111
>TC89387 similar to GP|17979249|gb|AAL49941.1 AT3g04610/F7O18_9 {Arabidopsis
thaliana}, partial (28%)
Length = 1205
Score = 28.1 bits (61), Expect = 4.6
Identities = 14/48 (29%), Positives = 28/48 (58%), Gaps = 3/48 (6%)
Frame = -3
Query: 49 EEVELHSYGAVENIPIYINAAFRASANNEAYLF---MNNEYVLINYAR 93
EE++ H + AVE I + + A + +++++L NE+V I Y++
Sbjct: 306 EEIQTHHHAAVEEIQTHHHGAVEETLSHQSHLLHRVHRNEFVEIPYSQ 163
>AW171653
Length = 390
Score = 27.7 bits (60), Expect = 6.0
Identities = 14/42 (33%), Positives = 22/42 (52%)
Frame = -1
Query: 182 AAFRAHSSNEAYLFRGGHYALINYSSKRLIHIETIRHGFPSL 223
AA R L G + I+ S ++ H+E++RHGF S+
Sbjct: 270 AAHRIFLQERFSLLAGVKHIRIHGQSPQVRHVESVRHGFGSV 145
>TC89930 similar to PIR|T47694|T47694 probable serine/threonine-specific
protein kinase - Arabidopsis thaliana, partial (53%)
Length = 1449
Score = 27.3 bits (59), Expect = 7.8
Identities = 14/42 (33%), Positives = 24/42 (56%)
Frame = +2
Query: 70 FRASANNEAYLFMNNEYVLINYARGSTNDYIINGPLYICDGY 111
+ A+ + + YLF ++EYVL R + +G LY+ +GY
Sbjct: 1103 YMANGSLDKYLFEDSEYVLSWEQRFKIIKGVASGLLYLHEGY 1228
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.139 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,401,217
Number of Sequences: 36976
Number of extensions: 130773
Number of successful extensions: 543
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 492
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 504
length of query: 313
length of database: 9,014,727
effective HSP length: 96
effective length of query: 217
effective length of database: 5,465,031
effective search space: 1185911727
effective search space used: 1185911727
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146590.2


