

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146590.14 - phase: 1 /pseudo
(299 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
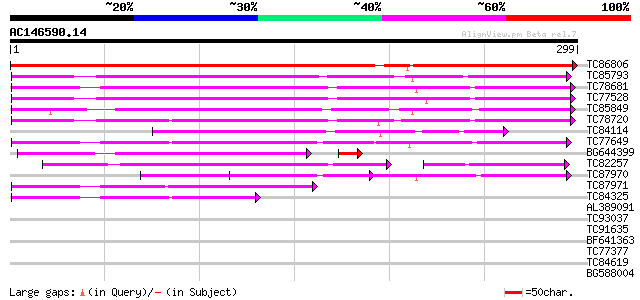
Score E
Sequences producing significant alignments: (bits) Value
TC86806 homologue to GP|3043430|emb|CAA06492.1 annexin {Cicer ar... 542 e-155
TC85793 similar to PIR|T31428|T31428 fiber annexin - upland cott... 174 5e-44
TC78681 similar to GP|3176098|emb|CAA75308.1 annexin {Medicago t... 169 1e-42
TC77528 similar to GP|3176098|emb|CAA75308.1 annexin {Medicago t... 160 4e-40
TC85849 homologue to PIR|T09552|T09552 annexin - alfalfa (fragme... 144 4e-35
TC78720 annexin; annexin [Medicago truncatula] 142 2e-34
TC84114 weakly similar to GP|3176098|emb|CAA75308.1 annexin {Med... 112 1e-25
TC77649 GP|3881978|emb|CAA72183.1 annexin-like protein {Medicago... 110 5e-25
BG644399 GP|8247363|emb annexin p34 {Solanum tuberosum}, partial... 100 3e-24
TC82257 62 4e-19
TC87970 similar to GP|3881978|emb|CAA72183.1 annexin-like protei... 84 9e-17
TC87971 similar to GP|3881978|emb|CAA72183.1 annexin-like protei... 70 1e-12
TC84325 similar to GP|3881978|emb|CAA72183.1 annexin-like protei... 43 2e-04
AL389091 homologue to GP|3881978|emb| annexin-like protein {Medi... 38 0.004
TC93037 weakly similar to GP|3004934|gb|AAC09237.1| annexin XIV ... 30 1.5
TC91635 similar to PIR|A84854|A84854 hypothetical protein At2g42... 29 1.9
BF641363 similar to GP|10567808|gb| putative glutathione S-trans... 27 9.5
TC77377 homologue to SP|P51850|DCP1_PEA Pyruvate decarboxylase i... 27 9.5
TC84619 27 9.5
BG588004 similar to GP|7242793|emb| cyclin A3.1 {Pisum sativum},... 27 9.5
>TC86806 homologue to GP|3043430|emb|CAA06492.1 annexin {Cicer arietinum},
complete
Length = 1236
Score = 542 bits (1396), Expect = e-155
Identities = 280/302 (92%), Positives = 285/302 (93%), Gaps = 3/302 (0%)
Frame = +2
Query: 1 HGVDEKSLIAVLGKWDPLERETYRKKTSHFFIEDHERQFQRWNDHCVRLLKHEFVRFKNA 60
HGVDEKSLIAVLGKWDPLERETYRKKTSHFFIEDHERQFQRWNDHCVRLLKHEFVRFKNA
Sbjct: 107 HGVDEKSLIAVLGKWDPLERETYRKKTSHFFIEDHERQFQRWNDHCVRLLKHEFVRFKNA 286
Query: 61 VVLWSMHPWERDARLAKEALKKGSISYGVLIEIACTRSSEELLGARKAYHSLFDHSIEED 120
VVLWSMHPWERDARLAKEALKKGSISYGVLIEIACTRSSEELLGARKAYHSLFDHSIEED
Sbjct: 287 VVLWSMHPWERDARLAKEALKKGSISYGVLIEIACTRSSEELLGARKAYHSLFDHSIEED 466
Query: 121 VASHIHGNDRKLLVALVSAYRYEGTKVKDDTAKSEAKTLSNAIKNAQNKPIVEDDEVIRI 180
VASHIHGNDRKLLVALVSAYRYEGTKVKDDTAKSEAKTLSNAIKNAQNKPIVEDDEVIRI
Sbjct: 467 VASHIHGNDRKLLVALVSAYRYEGTKVKDDTAKSEAKTLSNAIKNAQNKPIVEDDEVIRI 646
Query: 181 LATRSKLXLQAVYTAL*RDFWEES*RGI---INK*TLFKETVQCLCTPQVYFSKVLDAAL 237
LATRSKL LQAVY + + E S + + +N FKETVQCLCTPQVYFSKVLDAAL
Sbjct: 647 LATRSKLHLQAVY----KHYKEISGKNLEEDLND-LRFKETVQCLCTPQVYFSKVLDAAL 811
Query: 238 KNDVNKNIKKSLTRVIVTRADIDMKEIKAEYNNLYGVSLPQKIEETAKGNYKDFLLTLIA 297
KNDVNKNIKKSLTRVIVTRADIDMKEIKAEYNNLYGVSLPQKIEETAKGNYKDFLLTLIA
Sbjct: 812 KNDVNKNIKKSLTRVIVTRADIDMKEIKAEYNNLYGVSLPQKIEETAKGNYKDFLLTLIA 991
Query: 298 RG 299
RG
Sbjct: 992 RG 997
>TC85793 similar to PIR|T31428|T31428 fiber annexin - upland cotton,
complete
Length = 1272
Score = 174 bits (440), Expect = 5e-44
Identities = 115/306 (37%), Positives = 170/306 (54%), Gaps = 11/306 (3%)
Frame = +1
Query: 2 GVDEKSLIAVLGKWDPLERETYRKKTSHFFIEDHERQFQRWNDHCVRLLKHEFVR-FKNA 60
G +E +I++LG + ++R+ R+ + + ED ++ L E F+
Sbjct: 118 GTNENLIISILGHRNEVQRKVIREAYAKTYEED-----------LIKALNKELTSDFERL 264
Query: 61 VVLWSMHPWERDARLAKEALKKGSISYGVLIEIACTRSSEELLGARKAYHSLFDHSIEED 120
V LW++ ERDA LA EA K+ + S VL+E+ACTRSS++L A+KAYH+L S+EED
Sbjct: 265 VHLWTLESAERDAFLANEATKRWTSSNQVLVELACTRSSDQLFFAKKAYHALHKKSLEED 444
Query: 121 VASHIHGNDRKLLVALVSAYRYEGTKVKDDTAKSEAKTLSNAIKNAQNKPIVEDDEVIRI 180
VA H G+ RKLL+ LVS+YRYEG +V AK+EAK L I +K DD+ IRI
Sbjct: 445 VAYHTTGDFRKLLLPLVSSYRYEGDEVNLTIAKAEAKILHEKI----SKKAYNDDDFIRI 612
Query: 181 LATRSKLXLQAVYTAL*RDFWEES*RGIINK----------*TLFKETVQCLCTPQVYFS 230
LATRSK + A F ++ INK +L + TV+CL P+ YF+
Sbjct: 613 LATRSKAQINATLNHYKDAFGKD-----INKDLKEDPKNEYLSLLRSTVKCLVFPERYFA 777
Query: 231 KVLDAALKNDVNKNIKKSLTRVIVTRADIDMKEIKAEYNNLYGVSLPQKIEETAKGNYKD 290
K++ A+ + +LTRV+ TRA+ID+K I EY + L + I + G+Y+
Sbjct: 778 KIIREAINK--RGTDEGALTRVVATRAEIDLKIIAEEYQRRNSIPLDRAIVKDTTGDYEK 951
Query: 291 FLLTLI 296
LL ++
Sbjct: 952 MLLAIL 969
>TC78681 similar to GP|3176098|emb|CAA75308.1 annexin {Medicago truncatula},
partial (96%)
Length = 1254
Score = 169 bits (428), Expect = 1e-42
Identities = 109/302 (36%), Positives = 162/302 (53%), Gaps = 5/302 (1%)
Frame = +3
Query: 2 GVDEKSLIAVLGKWDPLERETYRKKTSHFFIEDHERQFQRWNDHCVRLLKHEFVRFKNAV 61
G DEKS+I +LG + +R+ RK + ED + RL F+ AV
Sbjct: 162 GTDEKSVITILGHRNVYQRQQIRKSYQEIYQEDILK----------RLESELSGDFERAV 311
Query: 62 VLWSMHPWERDARLAKEALKKGSISYGVLIEIACTRSSEELLGARKAYHSLFDHSIEEDV 121
W + P +RDA LA A+K GS SY V+IEI S EE+L R+AYH+ + HS+EED+
Sbjct: 312 YRWMLEPADRDAVLANVAIKDGSKSYHVIIEIVSVLSPEEVLAMRRAYHNRYKHSLEEDL 491
Query: 122 ASHIHGNDRKLLVALVSAYRYEGTKVKDDTAKSEAKTLSNAIKNAQNKPIVEDDEVIRIL 181
A+H G+ R+LLV LV+++RY G ++ AK+EA L +IK + +E IRIL
Sbjct: 492 AAHTTGHLRQLLVGLVTSFRYGGAEINPKLAKTEADILHESIKEKKG----NHEEAIRIL 659
Query: 182 ATRSKLXLQAVYTAL*RDFWEES*RGIINK*T-----LFKETVQCLCTPQVYFSKVLDAA 236
TRSK L A + D + +++ + T++C+ + Y+ K+L A
Sbjct: 660 TTRSKTQLLATFNRYRDDHGISITKKLLDNASDDFHKALHTTIRCINDHKKYYEKILRGA 839
Query: 237 LKNDVNKNIKKSLTRVIVTRADIDMKEIKAEYNNLYGVSLPQKIEETAKGNYKDFLLTLI 296
LK + LTRV+VTRA+ D+K+IK Y V L + + G+YK F+LTL+
Sbjct: 840 LKRVGTD--EDGLTRVVVTRAEKDLKDIKELYYKRNSVHLEDAVAKEISGDYKKFILTLL 1013
Query: 297 AR 298
+
Sbjct: 1014GK 1019
>TC77528 similar to GP|3176098|emb|CAA75308.1 annexin {Medicago truncatula},
partial (98%)
Length = 1378
Score = 160 bits (406), Expect = 4e-40
Identities = 105/303 (34%), Positives = 170/303 (55%), Gaps = 6/303 (1%)
Frame = +2
Query: 2 GVDEKSLIAVLGKWDPLERETYRKKTSHFFIEDHERQFQRWNDHCVRLLKHEFVR-FKNA 60
G D K++IA+LG + +R+ RK F ED ++ L+ E F+ A
Sbjct: 203 GADNKAIIAILGHRNVHQRQQIRKAYEELFEED-----------LIKRLESEISGDFERA 349
Query: 61 VVLWSMHPWERDARLAKEALKKGSISYGVLIEIACTRSSEELLGARKAYHSLFDHSIEED 120
V W + P +RDA L A++ G+ Y V+ EIA S+EELL R+AYH+ + SIEED
Sbjct: 350 VYRWMLDPADRDAVLINVAIRNGNKDYHVVAEIASVLSTEELLAVRRAYHNRYKRSIEED 529
Query: 121 VASHIHGNDRKLLVALVSAYRYEGTKVKDDTAKSEAKTLSNAIKNAQNKPIVEDDEVIRI 180
V++H G+ R+LLV LVS++RYEG ++ A++EA + ++K + ++EVIRI
Sbjct: 530 VSAHTTGHLRQLLVGLVSSFRYEGDEINAKLAQTEANIIHESVKEKKG----NNEEVIRI 697
Query: 181 LATRSKLXLQAVYTAL*RDFWEES*RGIINK*T-LFKET----VQCLCTPQVYFSKVLDA 235
L TRSK L A + + + ++++ + F++T ++C+ + Y+ KVL
Sbjct: 698 LTTRSKTQLVATFNRYRDEHGISISKKLLDQTSDDFQKTLHTAIRCINDHKKYYEKVLRN 877
Query: 236 ALKNDVNKNIKKSLTRVIVTRADIDMKEIKAEYNNLYGVSLPQKIEETAKGNYKDFLLTL 295
A+K + L+RVIVTRA+ D+++IK Y V L ++ + G+YK F+LTL
Sbjct: 878 AIKKFGTD--EDGLSRVIVTRAEKDLRDIKELYYKRNSVHLEDEVSKETSGDYKKFILTL 1051
Query: 296 IAR 298
+ +
Sbjct: 1052LGK 1060
>TC85849 homologue to PIR|T09552|T09552 annexin - alfalfa (fragment),
complete
Length = 1644
Score = 144 bits (363), Expect = 4e-35
Identities = 104/312 (33%), Positives = 161/312 (51%), Gaps = 15/312 (4%)
Frame = +3
Query: 2 GVDEKSLIAVLGKWDPLER----ETYRKKTSHFFIEDHERQFQRWNDHCVRLLKHEFVRF 57
G +E +I++L + +R ETY + ++D +++ F
Sbjct: 198 GTNEGLIISILAHRNAAQRKVIRETYTQTHGEDLLKDLDKELSS--------------DF 335
Query: 58 KNAVVLWSMHPWERDARLAKEALKKGSISYGVLIEIACTRSSEELLGARKAYHSLFDHSI 117
+ V+LW++ P ERDA LA +A K + + +++EIA TRS ELL A++AY + F S+
Sbjct: 336 EKVVLLWTLDPAERDAFLANQATKMLTSNNSIIVEIASTRSPLELLKAKQAYQARFKKSL 515
Query: 118 EEDVASHIHGNDRKLLVALVSAYRYEGTKVKDDTAKSEAKTLSNAIKNAQNKPIVEDDEV 177
EEDVA H + RKLLV LV +RYEG +V AKSEAK L I + D++
Sbjct: 516 EEDVAYHTSADIRKLLVPLVGIHRYEGDEVNMTLAKSEAKLLHEKIAD----KAYNHDDL 683
Query: 178 IRILATRSKLXLQAVYTAL*RDFWEES*RGIINK----------*TLFKETVQCLCTPQV 227
IRI+ TRSK L A +F +I+K L + ++ L P+
Sbjct: 684 IRIVTTRSKPQLNATLNHYNNEF-----GNVIDKDLDTDSDDEYLKLLRAAIKGLTYPEK 848
Query: 228 YFSKVLDAAL-KNDVNKNIKKSLTRVIVTRADIDMKEIKAEYNNLYGVSLPQKIEETAKG 286
YF ++L A+ K ++N +LTRV+ TRA++D++ I EY V L + I++ G
Sbjct: 849 YFEELLRLAINKMGTDEN---ALTRVVTTRAEVDLQRIAEEYQRRNSVPLDRAIDKDTSG 1019
Query: 287 NYKDFLLTLIAR 298
+Y+ LL L+ R
Sbjct: 1020DYQKILLALMGR 1055
>TC78720 annexin; annexin [Medicago truncatula]
Length = 1202
Score = 142 bits (357), Expect = 2e-34
Identities = 104/306 (33%), Positives = 157/306 (50%), Gaps = 9/306 (2%)
Frame = +2
Query: 2 GVDEKSLIAVLGKWDPLERETYRKKTSHFFIEDHERQFQRWNDHCVRLLKHEFVR-FKNA 60
G DEK++I +LG + + + RK + ED ++ L+ E F+ A
Sbjct: 110 GTDEKTVITILGHRNSNQIQQIRKAYEGIYNED-----------LIKRLESEIKGDFEKA 256
Query: 61 VVLWSMHPWERDARLAKEALKKGSISYGVLIEIACTRSSEELLGARKAYHSLFDHSIEED 120
V W + P ERDA LA A+K G +Y V++EI+ S EELL R+AY + HS+EED
Sbjct: 257 VYRWILEPAERDAVLANVAIKSGK-NYNVIVEISAVLSPEELLNVRRAYVKRYKHSLEED 433
Query: 121 VASHIHGNDRKLLVALVSAYRYEGTKVKDDTAKSEAKTLSNAIKNAQNKPIVEDDEVIRI 180
+A+H G+ R+LLV LV+A+RY G ++ A++EA L ++K + +E IRI
Sbjct: 434 LAAHTSGHLRQLLVGLVTAFRYVGDEINPKLAQTEAGILHESVKEKKG----SHEEAIRI 601
Query: 181 LATRSKLXLQAVY--------TAL*RDFWEES*RGIINK*TLFKETVQCLCTPQVYFSKV 232
L TRSK L A + T++ + +E G T++ Y+ KV
Sbjct: 602 LTTRSKTQLIATFNRYRETHGTSITKKLLDE---GSDEFQKALYTTIRSFNDHVKYYEKV 772
Query: 233 LDAALKNDVNKNIKKSLTRVIVTRADIDMKEIKAEYNNLYGVSLPQKIEETAKGNYKDFL 292
+ A+K + +LTRVIV+RA D+K I Y V L + + G+YK FL
Sbjct: 773 VRDAIKKVGTD--EDALTRVIVSRAQHDLKVISDVYYKRNSVLLEHVVAKETSGDYKKFL 946
Query: 293 LTLIAR 298
LTL+ +
Sbjct: 947 LTLLGK 964
>TC84114 weakly similar to GP|3176098|emb|CAA75308.1 annexin {Medicago
truncatula}, partial (55%)
Length = 562
Score = 112 bits (281), Expect = 1e-25
Identities = 73/196 (37%), Positives = 113/196 (57%), Gaps = 8/196 (4%)
Frame = +1
Query: 76 AKEALKKGSISYGVLIEIACTRSSEELLGARKAYHSLFDHSIEEDVASHIHGNDRKLLVA 135
A AL+ +I+Y +++EI+C S +EL R+AYH+ + S+EEDVA++ +G+ R+LLV
Sbjct: 1 ANIALRNANINYHLIVEISCVSSPDELFNLRRAYHNRYKRSLEEDVATNTNGHLRQLLVG 180
Query: 136 LVSAYRYEGTKVKDDTAKSEAKTLSNAIKNAQNKPIVEDDEVIRILATRSKLXLQAVYT- 194
LVS++RY+G++V A+ EA L AIKN +EVIRIL TRSK L A +
Sbjct: 181 LVSSFRYDGSEVNASLAQCEADMLHEAIKNKN----YNHEEVIRILTTRSKTQLVATFNC 348
Query: 195 -------AL*RDFWEES*RGIINK*TLFKETVQCLCTPQVYFSKVLDAALKNDVNKNIKK 247
A+ + +E G +L + C+ Y+ KVL A+ + +
Sbjct: 349 YRHDHGIAITKKLSDEGSDGFHKAVSL---AISCINDHNKYYEKVLRNAM--ETVGTDED 513
Query: 248 SLTRVIVTRADIDMKE 263
+LTRVIVTRA+ D+++
Sbjct: 514 ALTRVIVTRAEKDLED 561
>TC77649 GP|3881978|emb|CAA72183.1 annexin-like protein {Medicago sativa},
partial (96%)
Length = 1209
Score = 110 bits (276), Expect = 5e-25
Identities = 83/301 (27%), Positives = 145/301 (47%), Gaps = 6/301 (1%)
Frame = +1
Query: 2 GVDEKSLIAVLGKWDPLERETYRKKTSHFFIEDHERQFQRWNDHCVRLLKHEFVRFKNAV 61
G D ++I +L D +R +++ + E+ + RL+ + + AV
Sbjct: 121 GCDTTAVINILAHRDATQRAYLQQEYKATYSEELSK----------RLISELKGKLETAV 270
Query: 62 VLWSMHPWERDARLAKEALKKGSISYGVLIEIACTRSSEELLGARKAYHSLFDHSIEEDV 121
+LW P RDA + +++L + E+ C+R+ +L ++ YHS F +E ++
Sbjct: 271 LLWLPDPAARDAEIIRKSLVVDR-NLEAATEVICSRTPSQLQYLKQLYHSKFGVYLEHEI 447
Query: 122 ASHIHGNDRKLLVALVSAYRYEGTKVKDDTAKSEAKTLSNAIKNAQNKPIVEDDEVIRIL 181
+ G+ +K+L+ ++ R+EG +V + A+ +AK L A + K ++ ++I
Sbjct: 448 ELNTSGDHQKILLRYLTTPRHEGLEVNREIAQKDAKVLYKA---GEKKLGTDEKTFVQIF 618
Query: 182 ATRSKLXLQAVYTAL*RDFWEES*RGII------NK*TLFKETVQCLCTPQVYFSKVLDA 235
+ RS L AV ++ D + S + + N +C P YF+KVL
Sbjct: 619 SERSSAHLAAV-SSYYHDMYGHSLKKAVKNEASGNFGLALLTITECATNPAKYFAKVLYK 795
Query: 236 ALKNDVNKNIKKSLTRVIVTRADIDMKEIKAEYNNLYGVSLPQKIEETAKGNYKDFLLTL 295
A+K + +L RVIVTR +IDM+ IKAEY Y +L + GNY+DFLL L
Sbjct: 796 AMKGMGTND--STLIRVIVTRTEIDMQYIKAEYAKKYKKTLNDAVHSETSGNYRDFLLAL 969
Query: 296 I 296
+
Sbjct: 970 L 972
>BG644399 GP|8247363|emb annexin p34 {Solanum tuberosum}, partial (64%)
Length = 651
Score = 100 bits (250), Expect(2) = 3e-24
Identities = 63/156 (40%), Positives = 93/156 (59%), Gaps = 1/156 (0%)
Frame = +1
Query: 5 EKSLIAVLGKWDPLERETYRKKTSHFFIEDHERQFQRWNDHCVRLLKHEFVRFKNAVVLW 64
EK +I++L + +R+ R+ + F ED ++ R H F+ V++W
Sbjct: 91 EKLIISILAHRNAAQRKLIRQTYAETFGEDLLKELDRELTH----------DFEKLVLIW 240
Query: 65 SMHPWERDARLAKEALKKGSISYGVLIEIACTRSSEELLGARKAYHSLFDHSIEEDVASH 124
++ P ERDA LAKEA K+ + S VL+EIACTRS +EL+ AR+AYH+ S+EEDVA H
Sbjct: 241 TLDPSERDAYLAKEATKRWTKSNFVLVEIACTRSPKELVLAREAYHARNKKSLEEDVAYH 420
Query: 125 IH-GNDRKLLVALVSAYRYEGTKVKDDTAKSEAKTL 159
+ G+ + LVS+YRY G +V AK+E+K L
Sbjct: 421 TYWGSPQAFWYPLVSSYRYGGDEVDLRLAKAESKVL 528
Score = 28.1 bits (61), Expect(2) = 3e-24
Identities = 13/13 (100%), Positives = 13/13 (100%)
Frame = +2
Query: 174 DDEVIRILATRSK 186
DDEVIRILATRSK
Sbjct: 560 DDEVIRILATRSK 598
>TC82257
Length = 971
Score = 62.4 bits (150), Expect(2) = 4e-19
Identities = 44/185 (23%), Positives = 91/185 (48%), Gaps = 1/185 (0%)
Frame = +1
Query: 18 LERETYRKKTSHFFIEDHERQFQRWNDHCVRLLKHEFVRFKNAVVLWSMHPWERDARLAK 77
LER+ R+ + ED Q QR+++ +K + + LW + P +RDA +A+
Sbjct: 139 LERQQLRETYKTVYGEDLISQLQRYDEDGFSSMKF------STLSLWMLCPHDRDAFVAR 300
Query: 78 EALKKGSISYGVLIEIACTRSSEELLGARKAYHSLFDHSIEEDVASHIHGND-RKLLVAL 136
EAL++ ++ L+EI R S ++ +AYH +F +++D+ + + +K+L AL
Sbjct: 301 EALQQDETNFKALVEIFVGRKSSHIVLITQAYHKMFRRQLDQDIMNLDPPHPFQKILTAL 480
Query: 137 VSAYRYEGTKVKDDTAKSEAKTLSNAIKNAQNKPIVEDDEVIRILATRSKLXLQAVYTAL 196
++++ + +K +A+ L + + +++ V+ IL+ RS ++ L
Sbjct: 481 SASHKAHQADISHHISKCDARRLYETGEGSLG--AIDEAVVLEILSKRSISTIETNIFEL 654
Query: 197 *RDFW 201
W
Sbjct: 655 QTHLW 669
Score = 49.3 bits (116), Expect(2) = 4e-19
Identities = 29/78 (37%), Positives = 43/78 (54%), Gaps = 1/78 (1%)
Frame = +2
Query: 219 VQCLCTPQVYFSKVLDAALKNDVNKNIKKSLTRVIVTRADIDMKEIKAEYNNLYGVSLPQ 278
V C+C Y++K L ++K + K +L R +V+RADIDM EI+ + Y L
Sbjct: 737 VNCICNQAYYYAKELYTSIKRE--KRDIGTLARTLVSRADIDMDEIRRVFKEKYEKELGD 910
Query: 279 KI-EETAKGNYKDFLLTL 295
I E G+Y+DFL+ L
Sbjct: 911 FICESIPCGDYRDFLVAL 964
>TC87970 similar to GP|3881978|emb|CAA72183.1 annexin-like protein {Medicago
sativa}, partial (53%)
Length = 839
Score = 83.6 bits (205), Expect = 9e-17
Identities = 59/185 (31%), Positives = 97/185 (51%), Gaps = 5/185 (2%)
Frame = +1
Query: 117 IEEDVASHIHGNDRKLLVALVSAYRYEGTKVKDDTAKSEAKTLSNAIKNAQNKPIVEDDE 176
+E D+ + G+ +K+L+A VS R+EG +V + A+++AK L A + K ++
Sbjct: 7 LEHDMKRNASGDHKKILLAYVSTPRHEGPEVNREMAENDAKVLYKA---GEKKLGTDEKT 177
Query: 177 VIRILATRSKLXLQAVYTAL*RDFWEES*RGIINK*T-----LFKETVQCLCTPQVYFSK 231
++I + RS L A+ ++ + I N+ + VQC +P YF+K
Sbjct: 178 FVQIFSQRSAAQLAAINHFYHANYGHSLKKAIKNETSGNFAHALLTIVQCAESPAKYFAK 357
Query: 232 VLDAALKNDVNKNIKKSLTRVIVTRADIDMKEIKAEYNNLYGVSLPQKIEETAKGNYKDF 291
VL A+K + K L RVIVTR++ID+ IKAEY Y +L + G+Y+ F
Sbjct: 358 VLRKAMKGLGTDDTK--LMRVIVTRSEIDLHYIKAEYLKKYKKTLNDAVHSETSGHYRAF 531
Query: 292 LLTLI 296
LL+L+
Sbjct: 532 LLSLL 546
Score = 44.7 bits (104), Expect = 4e-05
Identities = 30/124 (24%), Positives = 57/124 (45%), Gaps = 1/124 (0%)
Frame = +1
Query: 70 ERDAR-LAKEALKKGSISYGVLIEIACTRSSEELLGARKAYHSLFDHSIEEDVASHIHGN 128
E DA+ L K KK ++I RS+ +L YH+ + HS+++ + + GN
Sbjct: 115 ENDAKVLYKAGEKKLGTDEKTFVQIFSQRSAAQLAAINHFYHANYGHSLKKAIKNETSGN 294
Query: 129 DRKLLVALVSAYRYEGTKVKDDTAKSEAKTLSNAIKNAQNKPIVEDDEVIRILATRSKLX 188
L+ +V A+S AK + ++ A +D +++R++ TRS++
Sbjct: 295 FAHALLTIVQC------------AESPAKYFAKVLRKAMKGLGTDDTKLMRVIVTRSEID 438
Query: 189 LQAV 192
L +
Sbjct: 439 LHYI 450
>TC87971 similar to GP|3881978|emb|CAA72183.1 annexin-like protein {Medicago
sativa}, partial (51%)
Length = 700
Score = 70.1 bits (170), Expect = 1e-12
Identities = 46/161 (28%), Positives = 82/161 (50%)
Frame = +2
Query: 2 GVDEKSLIAVLGKWDPLERETYRKKTSHFFIEDHERQFQRWNDHCVRLLKHEFVRFKNAV 61
G D ++I +L D +R +++ + ED + RL +F+NA+
Sbjct: 197 GCDTSAVINILAHRDATQRAYLQQEYRATYSEDLLK----------RLSSELSGKFENAI 346
Query: 62 VLWSMHPWERDARLAKEALKKGSISYGVLIEIACTRSSEELLGARKAYHSLFDHSIEEDV 121
+LW P RDA + K+ L S + E+ C+R+ +L R+ YH+ F ++ D+
Sbjct: 347 LLWMHDPATRDAIILKQTLTV-SKNLEATTEVICSRTPSQLQYLRQIYHTRFGVYLDHDI 523
Query: 122 ASHIHGNDRKLLVALVSAYRYEGTKVKDDTAKSEAKTLSNA 162
+ G+ +K+L+A VS R+EG +V + A+++AK L A
Sbjct: 524 ERNASGDHKKILLAYVSTPRHEGPEVNREMAENDAKGLYKA 646
>TC84325 similar to GP|3881978|emb|CAA72183.1 annexin-like protein {Medicago
sativa}, partial (57%)
Length = 771
Score = 42.7 bits (99), Expect = 2e-04
Identities = 31/132 (23%), Positives = 61/132 (45%), Gaps = 1/132 (0%)
Frame = +2
Query: 2 GVDEKSLIAVLGKWDPLERETYRKKTSHFFIEDHERQFQRWNDHCVRLLKHEFVRFKNAV 61
G D ++I +L D +R +++ + E+ + RL+ + + AV
Sbjct: 110 GCDTSAVINILAHRDATQRAYIQQEYRTTYAEELSK----------RLISELSGKLETAV 259
Query: 62 VLWSMHPWERDARLAKEALKKGSISYGVLIEIACTRSSEELLGARKAYHSLFDHSIEEDV 121
+LW P RDA + +++L + E+ C+R+ +L ++ YHS F +E ++
Sbjct: 260 LLWMPDPAGRDAEIIRKSLIVDK-NLEAATEVLCSRAPSQLQYLKQLYHSKFGVYLEHEI 436
Query: 122 AS-HIHGNDRKL 132
S H G+ + L
Sbjct: 437 ESNHFWGSSKDL 472
>AL389091 homologue to GP|3881978|emb| annexin-like protein {Medicago
sativa}, partial (22%)
Length = 489
Score = 38.1 bits (87), Expect = 0.004
Identities = 19/40 (47%), Positives = 24/40 (59%)
Frame = +3
Query: 248 SLTRVIVTRADIDMKEIKAEYNNLYGVSLPQKIEETAKGN 287
+L RVIVTR +IDM+ IKAEY Y +L + GN
Sbjct: 369 TLIRVIVTRTEIDMQYIKAEYAKKYKKTLNDAVHSETSGN 488
>TC93037 weakly similar to GP|3004934|gb|AAC09237.1| annexin XIV {Neurospora
crassa}, partial (33%)
Length = 778
Score = 29.6 bits (65), Expect = 1.5
Identities = 15/40 (37%), Positives = 22/40 (54%)
Frame = +3
Query: 2 GVDEKSLIAVLGKWDPLERETYRKKTSHFFIEDHERQFQR 41
G DEK+LI L DPL+ ET ++ F D E+ ++
Sbjct: 534 GTDEKALIRCLATKDPLQIETIKQAFKRNFNRDLEQDVKK 653
>TC91635 similar to PIR|A84854|A84854 hypothetical protein At2g42450
[imported] - Arabidopsis thaliana, partial (71%)
Length = 1919
Score = 29.3 bits (64), Expect = 1.9
Identities = 23/76 (30%), Positives = 36/76 (47%)
Frame = +1
Query: 96 TRSSEELLGARKAYHSLFDHSIEEDVASHIHGNDRKLLVALVSAYRYEGTKVKDDTAKSE 155
TR ++LL R Y + + + EE ++IH +L + A + E K+KD+ KS+
Sbjct: 37 TRYVKQLL--RNIYCTKYFLNREEPYRNYIHSVFNRLQLQRSRADKVEKDKLKDEQEKSQ 210
Query: 156 AKTLSNAIKNAQNKPI 171
SN K N I
Sbjct: 211 FSLKSNVKKADDNNGI 258
>BF641363 similar to GP|10567808|gb| putative glutathione S-transferase T3
{Lycopersicon esculentum}, partial (43%)
Length = 623
Score = 26.9 bits (58), Expect = 9.5
Identities = 19/63 (30%), Positives = 32/63 (50%), Gaps = 4/63 (6%)
Frame = -2
Query: 114 DHSIEEDVASHIHGNDRKLLVALVSAYRYEGTKV----KDDTAKSEAKTLSNAIKNAQNK 169
DH EED AS H + LV L+ YR+ +V +D TA++ N +++ +
Sbjct: 283 DHVEEEDFASRSHQYILESLVILIWTYRHAQEQVPSCKQDYTARAYHSCS*NLLQHIGIE 104
Query: 170 PIV 172
P++
Sbjct: 103 PLL 95
>TC77377 homologue to SP|P51850|DCP1_PEA Pyruvate decarboxylase isozyme 1 (EC
4.1.1.1) (PDC). [Garden pea] {Pisum sativum}, partial
(95%)
Length = 2128
Score = 26.9 bits (58), Expect = 9.5
Identities = 17/53 (32%), Positives = 22/53 (41%)
Frame = -2
Query: 224 TPQVYFSKVLDAALKNDVNKNIKKSLTRVIVTRADIDMKEIKAEYNNLYGVSL 276
T V+F LD A+K K K +VTR +K+ NN Y L
Sbjct: 1236 TAAVFFFTFLDNAVKKSAMKTQPKERPLPMVTRLGCTIKDFSFFINNEYPTEL 1078
>TC84619
Length = 590
Score = 26.9 bits (58), Expect = 9.5
Identities = 16/64 (25%), Positives = 30/64 (46%)
Frame = +2
Query: 117 IEEDVASHIHGNDRKLLVALVSAYRYEGTKVKDDTAKSEAKTLSNAIKNAQNKPIVEDDE 176
IE D++ I G+D + E KVKD+ ++ + K ++ P +EDD
Sbjct: 50 IENDISDSIKGHDE--------TSQAEDVKVKDNQEETPKLQDGSDEKTSEGTPKLEDDN 205
Query: 177 VIRI 180
+++
Sbjct: 206 KVKL 217
>BG588004 similar to GP|7242793|emb| cyclin A3.1 {Pisum sativum}, partial
(22%)
Length = 713
Score = 26.9 bits (58), Expect = 9.5
Identities = 18/67 (26%), Positives = 30/67 (43%)
Frame = +2
Query: 216 KETVQCLCTPQVYFSKVLDAALKNDVNKNIKKSLTRVIVTRADIDMKEIKAEYNNLYGVS 275
K+ V LC Q +V + NDVN N + V++ K + +N G
Sbjct: 320 KKAVAALCEQQRKRKRVPLGEITNDVNTNANANANDVVL---PTQKKRNVSTSSNSDGTP 490
Query: 276 LPQKIEE 282
+P+K+E+
Sbjct: 491 VPEKLEK 511
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.324 0.138 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,386,046
Number of Sequences: 36976
Number of extensions: 106768
Number of successful extensions: 754
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 720
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 725
length of query: 299
length of database: 9,014,727
effective HSP length: 96
effective length of query: 203
effective length of database: 5,465,031
effective search space: 1109401293
effective search space used: 1109401293
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146590.14


