

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146590.13 + phase: 0
(314 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
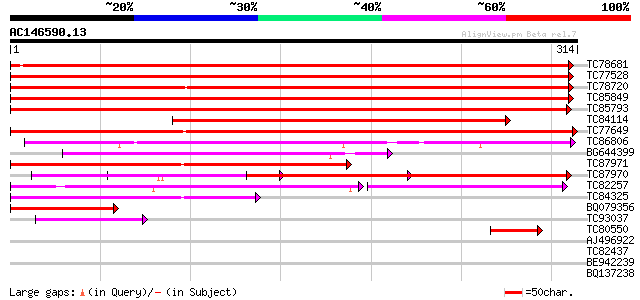
Score E
Sequences producing significant alignments: (bits) Value
TC78681 similar to GP|3176098|emb|CAA75308.1 annexin {Medicago t... 485 e-138
TC77528 similar to GP|3176098|emb|CAA75308.1 annexin {Medicago t... 463 e-131
TC78720 annexin; annexin [Medicago truncatula] 425 e-119
TC85849 homologue to PIR|T09552|T09552 annexin - alfalfa (fragme... 290 5e-79
TC85793 similar to PIR|T31428|T31428 fiber annexin - upland cott... 276 1e-74
TC84114 weakly similar to GP|3176098|emb|CAA75308.1 annexin {Med... 254 3e-68
TC77649 GP|3881978|emb|CAA72183.1 annexin-like protein {Medicago... 207 3e-54
TC86806 homologue to GP|3043430|emb|CAA06492.1 annexin {Cicer ar... 178 3e-45
BG644399 GP|8247363|emb annexin p34 {Solanum tuberosum}, partial... 135 3e-32
TC87971 similar to GP|3881978|emb|CAA72183.1 annexin-like protei... 134 6e-32
TC87970 similar to GP|3881978|emb|CAA72183.1 annexin-like protei... 132 2e-31
TC82257 71 2e-25
TC84325 similar to GP|3881978|emb|CAA72183.1 annexin-like protei... 97 1e-20
BQ079356 similar to GP|3881978|emb| annexin-like protein {Medica... 54 6e-08
TC93037 weakly similar to GP|3004934|gb|AAC09237.1| annexin XIV ... 44 8e-05
TC80550 40 9e-04
AJ496922 similar to GP|3176098|emb| annexin {Medicago truncatula... 30 1.6
TC82437 weakly similar to PIR|E71416|E71416 hypothetical protein... 28 4.6
BE942239 similar to PIR|A84809|A848 probable annexin [imported] ... 28 4.6
BQ137238 27 7.8
>TC78681 similar to GP|3176098|emb|CAA75308.1 annexin {Medicago truncatula},
partial (96%)
Length = 1254
Score = 485 bits (1249), Expect = e-138
Identities = 240/312 (76%), Positives = 276/312 (87%)
Frame = +3
Query: 1 MATLIAPMNHSPKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQEDL 60
MATL+ + SP EDA+ L KA KGWGTDE ++I I+G RN QRQQIR++YQ+IYQED+
Sbjct: 87 MATLVT-IKSSPVEDAEALQKAFKGWGTDEKSVITILGHRNVYQRQQIRKSYQEIYQEDI 263
Query: 61 IKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLAV 120
+KRLESELSG+FE+A+YRW+L+PADR AVLANVAIK +K YHVI+EI SVL P+E+LA+
Sbjct: 264 LKRLESELSGDFERAVYRWMLEPADRDAVLANVAIKDGSKSYHVIIEIVSVLSPEEVLAM 443
Query: 121 RHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVKN 180
R AYHNRYK+SLEED+AAHT+G+ RQLLVGLV+SFRY G EINP LAK EADILHE++K
Sbjct: 444 RRAYHNRYKHSLEEDLAAHTTGHLRQLLVGLVTSFRYGGAEINPKLAKTEADILHESIKE 623
Query: 181 KKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRCIDD 240
KKGN EE IRIL TRSKTQL ATFNRYRDDHG SI+KKLL+ ASDDF KA+H IRCI+D
Sbjct: 624 KKGNHEEAIRILTTRSKTQLLATFNRYRDDHGISITKKLLDNASDDFHKALHTTIRCIND 803
Query: 241 HKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKEISG 300
HKKYYEK+LRGALKR+GTDEDGLTRVV+TRAEKDLKDIKELYYKRNSVHLED VAKEISG
Sbjct: 804 HKKYYEKILRGALKRVGTDEDGLTRVVVTRAEKDLKDIKELYYKRNSVHLEDAVAKEISG 983
Query: 301 DYKKFLLTLLGK 312
DYKKF+LTLLGK
Sbjct: 984 DYKKFILTLLGK 1019
>TC77528 similar to GP|3176098|emb|CAA75308.1 annexin {Medicago truncatula},
partial (98%)
Length = 1378
Score = 463 bits (1192), Expect = e-131
Identities = 228/312 (73%), Positives = 270/312 (86%)
Frame = +2
Query: 1 MATLIAPMNHSPKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQEDL 60
MAT++ SP +DA+ L A KGWG D AIIAI+G RN QRQQIR+AY+++++EDL
Sbjct: 125 MATIVVHSQTSPVQDAEALRLAFKGWGADNKAIIAILGHRNVHQRQQIRKAYEELFEEDL 304
Query: 61 IKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLAV 120
IKRLESE+SG+FE+A+YRW+LDPADR AVL NVAI++ NKDYHV+ EIASVL +ELLAV
Sbjct: 305 IKRLESEISGDFERAVYRWMLDPADRDAVLINVAIRNGNKDYHVVAEIASVLSTEELLAV 484
Query: 121 RHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVKN 180
R AYHNRYK S+EEDV+AHT+G+ RQLLVGLVSSFRY+G EIN LA+ EA+I+HE+VK
Sbjct: 485 RRAYHNRYKRSIEEDVSAHTTGHLRQLLVGLVSSFRYEGDEINAKLAQTEANIIHESVKE 664
Query: 181 KKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRCIDD 240
KKGN EEVIRIL TRSKTQL ATFNRYRD+HG SISKKLL++ SDDF K +H AIRCI+D
Sbjct: 665 KKGNNEEVIRILTTRSKTQLVATFNRYRDEHGISISKKLLDQTSDDFQKTLHTAIRCIND 844
Query: 241 HKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKEISG 300
HKKYYEKVLR A+K+ GTDEDGL+RV++TRAEKDL+DIKELYYKRNSVHLED V+KE SG
Sbjct: 845 HKKYYEKVLRNAIKKFGTDEDGLSRVIVTRAEKDLRDIKELYYKRNSVHLEDEVSKETSG 1024
Query: 301 DYKKFLLTLLGK 312
DYKKF+LTLLGK
Sbjct: 1025DYKKFILTLLGK 1060
>TC78720 annexin; annexin [Medicago truncatula]
Length = 1202
Score = 425 bits (1092), Expect = e-119
Identities = 218/312 (69%), Positives = 260/312 (82%)
Frame = +2
Query: 1 MATLIAPMNHSPKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQEDL 60
MATL AP NHSP EDA+ L KA +GWGTDE +I I+G RN+ Q QQIR+AY+ IY EDL
Sbjct: 32 MATLSAPSNHSPNEDAEALRKAFEGWGTDEKTVITILGHRNSNQIQQIRKAYEGIYNEDL 211
Query: 61 IKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLAV 120
IKRLESE+ G+FEKA+YRWIL+PA+R AVLANVAIKS K+Y+VIVEI++VL P+ELL V
Sbjct: 212 IKRLESEIKGDFEKAVYRWILEPAERDAVLANVAIKS-GKNYNVIVEISAVLSPEELLNV 388
Query: 121 RHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVKN 180
R AY RYK+SLEED+AAHTSG+ RQLLVGLV++FRY G EINP LA+ EA ILHE+VK
Sbjct: 389 RRAYVKRYKHSLEEDLAAHTSGHLRQLLVGLVTAFRYVGDEINPKLAQTEAGILHESVKE 568
Query: 181 KKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRCIDD 240
KKG+ EE IRIL TRSKTQL ATFNRYR+ HG SI+KKLL+E SD+F KA++ IR +D
Sbjct: 569 KKGSHEEAIRILTTRSKTQLIATFNRYRETHGTSITKKLLDEGSDEFQKALYTTIRSFND 748
Query: 241 HKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKEISG 300
H KYYEKV+R A+K++GTDED LTRV+++RA+ DLK I ++YYKRNSV LE VAKE SG
Sbjct: 749 HVKYYEKVVRDAIKKVGTDEDALTRVIVSRAQHDLKVISDVYYKRNSVLLEHVVAKETSG 928
Query: 301 DYKKFLLTLLGK 312
DYKKFLLTLLGK
Sbjct: 929 DYKKFLLTLLGK 964
>TC85849 homologue to PIR|T09552|T09552 annexin - alfalfa (fragment),
complete
Length = 1644
Score = 290 bits (742), Expect = 5e-79
Identities = 157/313 (50%), Positives = 211/313 (67%), Gaps = 1/313 (0%)
Frame = +3
Query: 1 MATLIAPMN-HSPKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQED 59
MATL P N SP ED++ L A +GWGT+E II+I+ RNA QR+ IR+ Y + ED
Sbjct: 117 MATLKIPSNVPSPSEDSEQLRGAFQGWGTNEGLIISILAHRNAAQRKVIRETYTQTHGED 296
Query: 60 LIKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLA 119
L+K L+ ELS +FEK + W LDPA+R A LAN A K + + +IVEIAS P ELL
Sbjct: 297 LLKDLDKELSSDFEKVVLLWTLDPAERDAFLANQATKMLTSNNSIIVEIASTRSPLELLK 476
Query: 120 VRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVK 179
+ AY R+K SLEEDVA HTS R+LLV LV RY+G E+N LAK EA +LHE +
Sbjct: 477 AKQAYQARFKKSLEEDVAYHTSADIRKLLVPLVGIHRYEGDEVNMTLAKSEAKLLHEKIA 656
Query: 180 NKKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRCID 239
+K N +++IRI+ TRSK QL AT N Y ++ G I K L ++ D++LK + AI+ +
Sbjct: 657 DKAYNHDDLIRIVTTRSKPQLNATLNHYNNEFGNVIDKDLDTDSDDEYLKLLRAAIKGLT 836
Query: 240 DHKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKEIS 299
+KY+E++LR A+ ++GTDE+ LTRVV TRAE DL+ I E Y +RNSV L+ + K+ S
Sbjct: 837 YPEKYFEELLRLAINKMGTDENALTRVVTTRAEVDLQRIAEEYQRRNSVPLDRAIDKDTS 1016
Query: 300 GDYKKFLLTLLGK 312
GDY+K LL L+G+
Sbjct: 1017GDYQKILLALMGR 1055
>TC85793 similar to PIR|T31428|T31428 fiber annexin - upland cotton,
complete
Length = 1272
Score = 276 bits (705), Expect = 1e-74
Identities = 145/312 (46%), Positives = 203/312 (64%), Gaps = 1/312 (0%)
Frame = +1
Query: 1 MATLIAPMNHSP-KEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQED 59
M+TL P P +D + L KA GWGT+E+ II+I+G RN VQR+ IR+AY Y+ED
Sbjct: 37 MSTLSVPHPLPPVSDDVEQLRKAFSGWGTNENLIISILGHRNEVQRKVIREAYAKTYEED 216
Query: 60 LIKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLA 119
LIK L EL+ +FE+ ++ W L+ A+R A LAN A K V+VE+A +L
Sbjct: 217 LIKALNKELTSDFERLVHLWTLESAERDAFLANEATKRWTSSNQVLVELACTRSSDQLFF 396
Query: 120 VRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVK 179
+ AYH +K SLEEDVA HT+G R+LL+ LVSS+RY+G E+N +AK EA ILHE +
Sbjct: 397 AKKAYHALHKKSLEEDVAYHTTGDFRKLLLPLVSSYRYEGDEVNLTIAKAEAKILHEKIS 576
Query: 180 NKKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRCID 239
K N ++ IRIL TRSK Q+ AT N Y+D G I+K L + +++L + ++C+
Sbjct: 577 KKAYNDDDFIRILATRSKAQINATLNHYKDAFGKDINKDLKEDPKNEYLSLLRSTVKCLV 756
Query: 240 DHKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKEIS 299
++Y+ K++R A+ + GTDE LTRVV TRAE DLK I E Y +RNS+ L+ + K+ +
Sbjct: 757 FPERYFAKIIREAINKRGTDEGALTRVVATRAEIDLKIIAEEYQRRNSIPLDRAIVKDTT 936
Query: 300 GDYKKFLLTLLG 311
GDY+K LL +LG
Sbjct: 937 GDYEKMLLAILG 972
>TC84114 weakly similar to GP|3176098|emb|CAA75308.1 annexin {Medicago
truncatula}, partial (55%)
Length = 562
Score = 254 bits (649), Expect = 3e-68
Identities = 127/187 (67%), Positives = 153/187 (80%)
Frame = +1
Query: 91 ANVAIKSINKDYHVIVEIASVLQPQELLAVRHAYHNRYKNSLEEDVAAHTSGYHRQLLVG 150
AN+A+++ N +YH+IVEI+ V P EL +R AYHNRYK SLEEDVA +T+G+ RQLLVG
Sbjct: 1 ANIALRNANINYHLIVEISCVSSPDELFNLRRAYHNRYKRSLEEDVATNTNGHLRQLLVG 180
Query: 151 LVSSFRYDGVEINPILAKHEADILHEAVKNKKGNIEEVIRILITRSKTQLKATFNRYRDD 210
LVSSFRYDG E+N LA+ EAD+LHEA+KNK N EEVIRIL TRSKTQL ATFN YR D
Sbjct: 181 LVSSFRYDGSEVNASLAQCEADMLHEAIKNKNYNHEEVIRILTTRSKTQLVATFNCYRHD 360
Query: 211 HGFSISKKLLNEASDDFLKAVHVAIRCIDDHKKYYEKVLRGALKRIGTDEDGLTRVVITR 270
HG +I+KKL +E SD F KAV +AI CI+DH KYYEKVLR A++ +GTDED LTRV++TR
Sbjct: 361 HGIAITKKLSDEGSDGFHKAVSLAISCINDHNKYYEKVLRNAMETVGTDEDALTRVIVTR 540
Query: 271 AEKDLKD 277
AEKDL+D
Sbjct: 541 AEKDLED 561
>TC77649 GP|3881978|emb|CAA72183.1 annexin-like protein {Medicago sativa},
partial (96%)
Length = 1209
Score = 207 bits (528), Expect = 3e-54
Identities = 117/316 (37%), Positives = 190/316 (60%), Gaps = 2/316 (0%)
Frame = +1
Query: 1 MATLIAP-MNHSPKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQED 59
MATLI P + SP++DA L++A KG+G D +A+I I+ R+A QR ++Q Y+ Y E+
Sbjct: 40 MATLILPPIPPSPRDDAMQLYRAFKGFGCDTTAVINILAHRDATQRAYLQQEYKATYSEE 219
Query: 60 LIKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLA 119
L KRL SEL G E A+ W+ DPA R A + ++ ++++ E+ P +L
Sbjct: 220 LSKRLISELKGKLETAVLLWLPDPAARDAEIIRKSLV-VDRNLEAATEVICSRTPSQLQY 396
Query: 120 VRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVK 179
++ YH+++ LE ++ +TSG H+++L+ +++ R++G+E+N +A+ +A +L++A +
Sbjct: 397 LKQLYHSKFGVYLEHEIELNTSGDHQKILLRYLTTPRHEGLEVNREIAQKDAKVLYKAGE 576
Query: 180 NKKGNIEEV-IRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRCI 238
K G E+ ++I RS L A + Y D +G S+ K + NEAS +F A+ C
Sbjct: 577 KKLGTDEKTFVQIFSERSSAHLAAVSSYYHDMYGHSLKKAVKNEASGNFGLALLTITECA 756
Query: 239 DDHKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKEI 298
+ KY+ KVL A+K +GT++ L RV++TR E D++ IK Y K+ L D V E
Sbjct: 757 TNPAKYFAKVLYKAMKGMGTNDSTLIRVIVTRTEIDMQYIKAEYAKKYKKTLNDAVHSET 936
Query: 299 SGDYKKFLLTLLGKGH 314
SG+Y+ FLL LLG H
Sbjct: 937 SGNYRDFLLALLGPNH 984
>TC86806 homologue to GP|3043430|emb|CAA06492.1 annexin {Cicer arietinum},
complete
Length = 1236
Score = 178 bits (451), Expect = 3e-45
Identities = 113/322 (35%), Positives = 177/322 (54%), Gaps = 17/322 (5%)
Frame = +2
Query: 9 NHSPKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQED--------- 59
N + ++ + + +A G G DE ++IA++G+ + ++R+ R+ + ED
Sbjct: 56 NMAFNQELEAITQAFSGHGVDEKSLIAVLGKWDPLERETYRKKTSHFFIEDHERQFQRWN 235
Query: 60 --LIKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQEL 117
++ L+ E F+ A+ W + P +R A LA A+K + Y V++EIA +EL
Sbjct: 236 DHCVRLLKHEFV-RFKNAVVLWSMHPWERDARLAKEALKKGSISYGVLIEIACTRSSEEL 412
Query: 118 LAVRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEA 177
L R AYH+ + +S+EEDVA+H G R+LLV LVS++RY+G ++ AK EA L A
Sbjct: 413 LGARKAYHSLFDHSIEEDVASHIHGNDRKLLVALVSAYRYEGTKVKDDTAKSEAKTLSNA 592
Query: 178 VKNKKG----NIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHV 233
+KN + +EVIRIL TRSK L+A + Y++ IS K L E +D
Sbjct: 593 IKNAQNKPIVEDDEVIRILATRSKLHLQAVYKHYKE-----ISGKNLEEDLNDL--RFKE 751
Query: 234 AIRCIDDHKKYYEKVLRGALKRIGTD--EDGLTRVVITRAEKDLKDIKELYYKRNSVHLE 291
++C+ + Y+ KVL ALK + LTRV++TRA+ D+K+IK Y V L
Sbjct: 752 TVQCLCTPQVYFSKVLDAALKNDVNKNIKKSLTRVIVTRADIDMKEIKAEYNNLYGVSLP 931
Query: 292 DTVAKEISGDYKKFLLTLLGKG 313
+ + G+YK FLLTL+ +G
Sbjct: 932 QKIEETAKGNYKDFLLTLIARG 997
>BG644399 GP|8247363|emb annexin p34 {Solanum tuberosum}, partial (64%)
Length = 651
Score = 135 bits (339), Expect = 3e-32
Identities = 82/189 (43%), Positives = 112/189 (58%), Gaps = 6/189 (3%)
Frame = +1
Query: 30 ESAIIAIMGQRNAVQRQQIRQAYQDIYQEDLIKRLESELSGNFEKAMYRWILDPADRYAV 89
E II+I+ RNA QR+ IRQ Y + + EDL+K L+ EL+ +FEK + W LDP++R A
Sbjct: 91 EKLIISILAHRNAAQRKLIRQTYAETFGEDLLKELDRELTHDFEKLVLIWTLDPSERDAY 270
Query: 90 LANVAIKSINKDYHVIVEIASVLQPQELLAVRHAYHNRYKNSLEEDVAAHTS-GYHRQLL 148
LA A K K V+VEIA P+EL+ R AYH R K SLEEDVA HT G +
Sbjct: 271 LAKEATKRWTKSNFVLVEIACTRSPKELVLAREAYHARNKKSLEEDVAYHTYWGSPQAFW 450
Query: 149 VGLVSSFRYDGVEINPILAKHEADILHE-----AVKNKKGNIEEVIRILITRSKTQLKAT 203
LVSS+RY G E++ LAK E+ +LHE ++ + G+ + + QL AT
Sbjct: 451 YPLVSSYRYGGDEVDLRLAKAESKVLHEKDLR*GLQ*R*GH*N-----FSHKEQAQLNAT 615
Query: 204 FNRYRDDHG 212
N Y+D++G
Sbjct: 616 LNHYQDEYG 642
>TC87971 similar to GP|3881978|emb|CAA72183.1 annexin-like protein {Medicago
sativa}, partial (51%)
Length = 700
Score = 134 bits (336), Expect = 6e-32
Identities = 71/190 (37%), Positives = 120/190 (62%), Gaps = 1/190 (0%)
Frame = +2
Query: 1 MATL-IAPMNHSPKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQED 59
M+TL + P+ SP++DA L +A KG+G D SA+I I+ R+A QR ++Q Y+ Y ED
Sbjct: 116 MSTLNVPPIPPSPRDDAMQLHRAFKGFGCDTSAVINILAHRDATQRAYLQQEYRATYSED 295
Query: 60 LIKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLA 119
L+KRL SELSG FE A+ W+ DPA R A++ + +++K+ E+ P +L
Sbjct: 296 LLKRLSSELSGKFENAILLWMHDPATRDAIILKQTL-TVSKNLEATTEVICSRTPSQLQY 472
Query: 120 VRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVK 179
+R YH R+ L+ D+ + SG H+++L+ VS+ R++G E+N +A+++A L++A +
Sbjct: 473 LRQIYHTRFGVYLDHDIERNASGDHKKILLAYVSTPRHEGPEVNREMAENDAKGLYKAGE 652
Query: 180 NKKGNIEEVI 189
K G + +++
Sbjct: 653 XKLGLMXKLL 682
>TC87970 similar to GP|3881978|emb|CAA72183.1 annexin-like protein {Medicago
sativa}, partial (53%)
Length = 839
Score = 132 bits (332), Expect = 2e-31
Identities = 72/181 (39%), Positives = 113/181 (61%), Gaps = 1/181 (0%)
Frame = +1
Query: 132 LEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVKNKKGNIEEV-IR 190
LE D+ + SG H+++L+ VS+ R++G E+N +A+++A +L++A + K G E+ ++
Sbjct: 7 LEHDMKRNASGDHKKILLAYVSTPRHEGPEVNREMAENDAKVLYKAGEKKLGTDEKTFVQ 186
Query: 191 ILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRCIDDHKKYYEKVLR 250
I RS QL A + Y ++G S+ K + NE S +F A+ ++C + KY+ KVLR
Sbjct: 187 IFSQRSAAQLAAINHFYHANYGHSLKKAIKNETSGNFAHALLTIVQCAESPAKYFAKVLR 366
Query: 251 GALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKEISGDYKKFLLTLL 310
A+K +GTD+ L RV++TR+E DL IK Y K+ L D V E SG Y+ FLL+LL
Sbjct: 367 KAMKGLGTDDTKLMRVIVTRSEIDLHYIKAEYLKKYKKTLNDAVHSETSGHYRAFLLSLL 546
Query: 311 G 311
G
Sbjct: 547 G 549
Score = 70.1 bits (170), Expect = 1e-12
Identities = 45/144 (31%), Positives = 72/144 (49%), Gaps = 4/144 (2%)
Frame = +1
Query: 13 KEDADVLWKA-VKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQEDLIKRLESELSGN 71
+ DA VL+KA K GTDE + I QR+A Q I Y Y L K +++E SGN
Sbjct: 115 ENDAKVLYKAGEKKLGTDEKTFVQIFSQRSAAQLAAINHFYHANYGHSLKKAIKNETSGN 294
Query: 72 FEKAMYRWIL---DPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLAVRHAYHNRY 128
F A+ + PA +A + A+K + D ++ + +L ++ Y +Y
Sbjct: 295 FAHALLTIVQCAESPAKYFAKVLRKAMKGLGTDDTKLMRVIVTRSEIDLHYIKAEYLKKY 474
Query: 129 KNSLEEDVAAHTSGYHRQLLVGLV 152
K +L + V + TSG++R L+ L+
Sbjct: 475 KKTLNDAVHSETSGHYRAFLLSLL 546
Score = 49.7 bits (117), Expect = 1e-06
Identities = 47/181 (25%), Positives = 79/181 (42%), Gaps = 12/181 (6%)
Frame = +1
Query: 55 IYQEDLIKRLESELSGNFEKAMYRWILDP-----------ADRYA-VLANVAIKSINKDY 102
+Y E +KR SG+ +K + ++ P A+ A VL K + D
Sbjct: 1 VYLEHDMKR---NASGDHKKILLAYVSTPRHEGPEVNREMAENDAKVLYKAGEKKLGTDE 171
Query: 103 HVIVEIASVLQPQELLAVRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEI 162
V+I S +L A+ H YH Y +SL++ + TSG L+ +V
Sbjct: 172 KTFVQIFSQRSAAQLAAINHFYHANYGHSLKKAIKNETSGNFAHALLTIVQCAESP---- 339
Query: 163 NPILAKHEADILHEAVKNKKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNE 222
AK+ A +L +A+K + +++R+++TRS+ L Y + KK LN+
Sbjct: 340 ----AKYFAKVLRKAMKGLGTDDTKLMRVIVTRSEIDLHYIKAEYLKKY-----KKTLND 492
Query: 223 A 223
A
Sbjct: 493 A 495
>TC82257
Length = 971
Score = 70.9 bits (172), Expect(2) = 2e-25
Identities = 53/204 (25%), Positives = 102/204 (49%), Gaps = 8/204 (3%)
Frame = +1
Query: 1 MATLIAPM-NHSPKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQED 59
+AT I M NH+ + D + ++ G S ++ + ++RQQ+R+ Y+ +Y ED
Sbjct: 22 IATNILTMTNHNFEHDCKKIHDSLGGL----SQLVPSLSCLTILERQQLRETYKTVYGED 189
Query: 60 LIKRLESELSGNFEKAMYR----WILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQ 115
LI +L+ F + W+L P DR A +A A++ ++ +VEI +
Sbjct: 190 LISQLQRYDEDGFSSMKFSTLSLWMLCPHDRDAFVAREALQQDETNFKALVEIFVGRKSS 369
Query: 116 ELLAVRHAYHNRYKNSLEEDVAAHTSGY-HRQLLVGLVSSFRYDGVEINPILAKHEADIL 174
++ + AYH ++ L++D+ + +++L L +S + +I+ ++K +A L
Sbjct: 370 HIVLITQAYHKMFRRQLDQDIMNLDPPHPFQKILTALSASHKAHQADISHHISKCDARRL 549
Query: 175 HEAVKNKKGNIEE--VIRILITRS 196
+E + G I+E V+ IL RS
Sbjct: 550 YETGEGSLGAIDEAVVLEILSKRS 621
Score = 62.0 bits (149), Expect(2) = 2e-25
Identities = 35/112 (31%), Positives = 60/112 (53%), Gaps = 1/112 (0%)
Frame = +2
Query: 199 QLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRCIDDHKKYYEKVLRGALKRIGT 258
QLK TF Y+ +G +K + F K++ V + CI + YY K L ++KR
Sbjct: 629 QLKLTFLSYKHIYGHDYTKSIKRGNYGQFGKSLMVVVNCICNQAYYYAKELYTSIKREKR 808
Query: 259 DEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKEI-SGDYKKFLLTL 309
D L R +++RA+ D+ +I+ ++ ++ L D + + I GDY+ FL+ L
Sbjct: 809 DIGTLARTLVSRADIDMDEIRRVFKEKYEKELGDFICESIPCGDYRDFLVAL 964
>TC84325 similar to GP|3881978|emb|CAA72183.1 annexin-like protein {Medicago
sativa}, partial (57%)
Length = 771
Score = 96.7 bits (239), Expect = 1e-20
Identities = 53/140 (37%), Positives = 85/140 (59%), Gaps = 1/140 (0%)
Frame = +2
Query: 1 MATLIAP-MNHSPKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQED 59
MATL+ P + SP++DA L++A KG+G D SA+I I+ R+A QR I+Q Y+ Y E+
Sbjct: 29 MATLVVPPIPPSPRDDAMQLYRAFKGFGCDTSAVINILAHRDATQRAYIQQEYRTTYAEE 208
Query: 60 LIKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLA 119
L KRL SELSG E A+ W+ DPA R A + ++ ++K+ E+ P +L
Sbjct: 209 LSKRLISELSGKLETAVLLWMPDPAGRDAEIIRKSL-IVDKNLEAATEVLCSRAPSQLQY 385
Query: 120 VRHAYHNRYKNSLEEDVAAH 139
++ YH+++ LE ++ ++
Sbjct: 386 LKQLYHSKFGVYLEHEIESN 445
>BQ079356 similar to GP|3881978|emb| annexin-like protein {Medicago sativa},
partial (18%)
Length = 489
Score = 54.3 bits (129), Expect = 6e-08
Identities = 28/61 (45%), Positives = 41/61 (66%), Gaps = 1/61 (1%)
Frame = +1
Query: 1 MATL-IAPMNHSPKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQED 59
M+TL + P+ SP++DA L +A KG+G D SA+I I+ R+A QR ++Q Y+ Y ED
Sbjct: 115 MSTLNVPPIPPSPRDDAIQLHRAFKGFGCDTSAVINILAHRDATQRAYLQQEYRATYSED 294
Query: 60 L 60
L
Sbjct: 295 L 297
>TC93037 weakly similar to GP|3004934|gb|AAC09237.1| annexin XIV {Neurospora
crassa}, partial (33%)
Length = 778
Score = 43.9 bits (102), Expect = 8e-05
Identities = 22/62 (35%), Positives = 37/62 (59%)
Frame = +3
Query: 15 DADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQEDLIKRLESELSGNFEK 74
DA A+KG+GTDE A+I + ++ +Q + I+QA++ + DL + ++ E S F K
Sbjct: 498 DATAARNAMKGFGTDEKALIRCLATKDPLQIETIKQAFKRNFNRDLEQDVKKETSSWFGK 677
Query: 75 AM 76
M
Sbjct: 678 GM 683
>TC80550
Length = 522
Score = 40.4 bits (93), Expect = 9e-04
Identities = 18/29 (62%), Positives = 23/29 (79%)
Frame = +2
Query: 267 VITRAEKDLKDIKELYYKRNSVHLEDTVA 295
++ R +L+DIKEL YKRNSVHLED V+
Sbjct: 167 LLCRGADNLRDIKELSYKRNSVHLEDAVS 253
>AJ496922 similar to GP|3176098|emb| annexin {Medicago truncatula}, partial
(5%)
Length = 132
Score = 29.6 bits (65), Expect = 1.6
Identities = 18/38 (47%), Positives = 23/38 (60%), Gaps = 1/38 (2%)
Frame = +1
Query: 191 ILITRSKTQLKATFNRYRDDHGFSIS-KKLLNEASDDF 227
IL TR +TQL ATFNRY + + ++L SDDF
Sbjct: 16 ILTTRKQTQLVATFNRY*GLKTWHLH**EMLGSTSDDF 129
>TC82437 weakly similar to PIR|E71416|E71416 hypothetical protein -
Arabidopsis thaliana, partial (42%)
Length = 1680
Score = 28.1 bits (61), Expect = 4.6
Identities = 22/91 (24%), Positives = 40/91 (43%)
Frame = +1
Query: 89 VLANVAIKSINKDYHVIVEIASVLQPQELLAVRHAYHNRYKNSLEEDVAAHTSGYHRQLL 148
V VA+ N ++V + +V P+ L+ ++Y + L + AH+ Y + L
Sbjct: 808 VAETVALNHSNVKWYVFGDDDTVFFPENLVKTL----SKYDHELWYYIGAHSEIYEQNRL 975
Query: 149 VGLVSSFRYDGVEINPILAKHEADILHEAVK 179
G +F G I+ LAK A + ++
Sbjct: 976 FGFGMAFGGAGFAISSSLAKVLAKVFDSCLE 1068
>BE942239 similar to PIR|A84809|A848 probable annexin [imported] -
Arabidopsis thaliana, partial (6%)
Length = 545
Score = 28.1 bits (61), Expect = 4.6
Identities = 13/40 (32%), Positives = 23/40 (57%)
Frame = +2
Query: 229 KAVHVAIRCIDDHKKYYEKVLRGALKRIGTDEDGLTRVVI 268
K + ++ R HK+ V+R ++ +GTDED L R ++
Sbjct: 437 KMIAISFRFTSMHKQ----VIRDSIVGLGTDEDSLNRAIV 544
>BQ137238
Length = 1023
Score = 27.3 bits (59), Expect = 7.8
Identities = 12/39 (30%), Positives = 20/39 (50%)
Frame = +3
Query: 118 LAVRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFR 156
L + HA+H+ E V +HT H + L+S++R
Sbjct: 804 LTLTHAHHHSTTTHTETAVRSHTIATHTMNIAMLISAYR 920
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.135 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,003,033
Number of Sequences: 36976
Number of extensions: 89236
Number of successful extensions: 455
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 436
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 439
length of query: 314
length of database: 9,014,727
effective HSP length: 96
effective length of query: 218
effective length of database: 5,465,031
effective search space: 1191376758
effective search space used: 1191376758
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146590.13


