

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146588.9 + phase: 0
(373 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
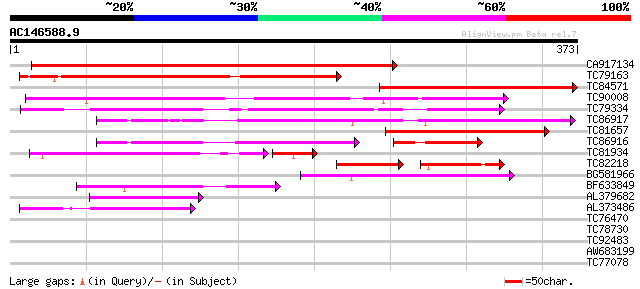
Score E
Sequences producing significant alignments: (bits) Value
CA917134 weakly similar to PIR|E96724|E96 hypothetical protein F... 405 e-113
TC79163 weakly similar to PIR|E96724|E96724 hypothetical protein... 265 2e-71
TC84571 weakly similar to SP|P29136|MEP1_SOYBN Metalloendoprotei... 264 4e-71
TC90008 weakly similar to GP|4249376|gb|AAD14473.1| Strong simil... 192 1e-49
TC79334 MtN9 166 2e-41
TC86917 similar to GP|3776080|emb|CAA77093.1 MtN9 {Medicago trun... 147 6e-36
TC81657 weakly similar to PIR|T51957|T51957 metalloproteinase (E... 144 7e-35
TC86916 similar to GP|3776080|emb|CAA77093.1 MtN9 {Medicago trun... 101 3e-31
TC81934 weakly similar to PIR|E71433|E71433 probable metalloprot... 96 3e-23
TC82218 similar to GP|7159629|emb|CAB76364.1 matrix metalloprote... 60 1e-18
BG581966 similar to GP|3776080|emb MtN9 {Medicago truncatula}, p... 90 2e-18
BF633849 weakly similar to PIR|E71433|E714 probable metalloprote... 79 3e-15
AL379682 weakly similar to SP|P29136|MEP1_ Metalloendoproteinase... 74 1e-13
AL373486 weakly similar to GP|16901508|gb| matrix metalloprotein... 67 8e-12
TC76470 similar to PIR|D84651|D84651 hypothetical protein At2g25... 29 2.6
TC78730 similar to GP|13876520|gb|AAK43496.1 hypothetical protei... 29 3.3
TC92483 similar to GP|21592537|gb|AAM64486.1 unknown {Arabidopsi... 28 5.7
AW683199 similar to PIR|B85043|B85 probable GH3-like protein [im... 28 5.7
TC77078 similar to GP|21537078|gb|AAM61419.1 putaive DNA-binding... 28 7.4
>CA917134 weakly similar to PIR|E96724|E96 hypothetical protein F20P5.11
[imported] - Arabidopsis thaliana, partial (26%)
Length = 723
Score = 405 bits (1041), Expect = e-113
Identities = 200/241 (82%), Positives = 204/241 (83%)
Frame = +1
Query: 15 SLTLLIVSARLFPDVPSWIPPGTLPVGPWDAFRNFTGCRQGENYNGLSNLKNYFQHFGYI 74
SLTLLIVSARLFPDVPSWIPPGTLPVGPWDAFRNFTGCRQGENYNGLSNLKNYFQHFGYI
Sbjct: 1 SLTLLIVSARLFPDVPSWIPPGTLPVGPWDAFRNFTGCRQGENYNGLSNLKNYFQHFGYI 180
Query: 75 PRSPKSNFSDDFDDDLQEAIKTYQKNFNLNVTGELDDMTLRQVMLPRCGVADIINGTTTM 134
PRSPKSNFSDDFDDDLQEAIKTYQKNFNLNVTGELDDMTLRQVMLPRCGVADIINGTTTM
Sbjct: 181 PRSPKSNFSDDFDDDLQEAIKTYQKNFNLNVTGELDDMTLRQVMLPRCGVADIINGTTTM 360
Query: 135 NAGKDTETTSNSDSKLRFHTVSHFTVFPGQPRWPEGKQELTYAFFPGNELTETVKSVFAT 194
NAGKDTETTSNSDSKLRFHTVSHFTVFPGQPRWPEGKQELTYAFFPGNELTETVKSVFAT
Sbjct: 361 NAGKDTETTSNSDSKLRFHTVSHFTVFPGQPRWPEGKQELTYAFFPGNELTETVKSVFAT 540
Query: 195 AFARWSEVTTLKFTETTLYSGADIKIGFFNGDHGDGEPFDGSLGTLAHAFSPRNGRFHLD 254
AFARWSEVTTL + ++ FF + GRFHLD
Sbjct: 541 AFARWSEVTTLNSPKQRYIPALILRSDFFTATMETASRLTEAWER*RMLSLKGTGRFHLD 720
Query: 255 A 255
A
Sbjct: 721 A 723
>TC79163 weakly similar to PIR|E96724|E96724 hypothetical protein F20P5.11
[imported] - Arabidopsis thaliana, partial (32%)
Length = 762
Score = 265 bits (677), Expect = 2e-71
Identities = 126/215 (58%), Positives = 158/215 (72%), Gaps = 3/215 (1%)
Frame = +2
Query: 7 IFPLTISFSLTLLIVSARLFPD---VPSWIPPGTLPVGPWDAFRNFTGCRQGENYNGLSN 63
IF +T+ S+TL VSAR FPD +P+W P G WD ++NFTGC +G+ Y+GLS
Sbjct: 140 IFYITL-ISITLTSVSARFFPDPSSMPAW-NSSNAPPGAWDGYKNFTGCSRGKTYDGLSK 313
Query: 64 LKNYFQHFGYIPRSPKSNFSDDFDDDLQEAIKTYQKNFNLNVTGELDDMTLRQVMLPRCG 123
LKNYF HFGYIP P SNF+DDFD+ L+ A++TYQKNFNLN+TGELDD T+ ++ PRCG
Sbjct: 314 LKNYFNHFGYIPNGPPSNFTDDFDEALESAVRTYQKNFNLNITGELDDATMNYIVKPRCG 493
Query: 124 VADIINGTTTMNAGKDTETTSNSDSKLRFHTVSHFTVFPGQPRWPEGKQELTYAFFPGNE 183
VADIINGTT+MN+GK +++N FHTV+H++ FPGQPRWPEG Q LTYAF P
Sbjct: 494 VADIINGTTSMNSGKFNSSSTN------FHTVAHYSFFPGQPRWPEGTQTLTYAFDPSEN 655
Query: 184 LTETVKSVFATAFARWSEVTTLKFTETTLYSGADI 218
L + K VFA AF +WS+VTT+ FTE T YS +DI
Sbjct: 656 LDDATKQVFANAFNQWSKVTTITFTEATSYSSSDI 760
>TC84571 weakly similar to SP|P29136|MEP1_SOYBN Metalloendoproteinase 1
precursor (EC 3.4.24.-) (SMEP1). [Soybean] {Glycine
max}, partial (28%)
Length = 506
Score = 264 bits (675), Expect = 4e-71
Identities = 130/130 (100%), Positives = 130/130 (100%)
Frame = +2
Query: 244 FSPRNGRFHLDAAEDWVVSGDVSKSSLPTAVDLESVAVHEIGHLLGLGHSSEEEAIMFPT 303
FSPRNGRFHLDAAEDWVVSGDVSKSSLPTAVDLESVAVHEIGHLLGLGHSSEEEAIMFPT
Sbjct: 2 FSPRNGRFHLDAAEDWVVSGDVSKSSLPTAVDLESVAVHEIGHLLGLGHSSEEEAIMFPT 181
Query: 304 ISSRMKKVVLADDDVRGIQYLYGTNPSFNGSTVISSPERNIGNGGCSLTSLWSPWRLFSL 363
ISSRMKKVVLADDDVRGIQYLYGTNPSFNGSTVISSPERNIGNGGCSLTSLWSPWRLFSL
Sbjct: 182 ISSRMKKVVLADDDVRGIQYLYGTNPSFNGSTVISSPERNIGNGGCSLTSLWSPWRLFSL 361
Query: 364 LTFVLSHFLL 373
LTFVLSHFLL
Sbjct: 362 LTFVLSHFLL 391
>TC90008 weakly similar to GP|4249376|gb|AAD14473.1| Strong similarity to
GP|2829864 F3I6.6 zinc metalloproteinase homolog from
Arabidopsis thaliana BAC, partial (21%)
Length = 1173
Score = 192 bits (489), Expect = 1e-49
Identities = 123/325 (37%), Positives = 170/325 (51%), Gaps = 7/325 (2%)
Frame = +2
Query: 11 TISFSLTLLIVSARLFPDVPSWIPPGTLPVGPWDAFRNF--TGCRQGENYNGLSNLKNYF 68
+ISF+ + ++ P I L W+ ++ + G+ +G+S+LK Y
Sbjct: 116 SISFAPAIGLIVRSRIPFYLKRITSSKLFSSIWNKLKSIFNNALKIGDIASGISDLKEYL 295
Query: 69 QHFGYIPRSP-KSNFSDDFDDDLQEAIKTYQKNFNLNVTGELDDMTLRQVMLPRCGVADI 127
Q FGY+ S SNF+D F ++LQ AI +Q NFNLN TG+LD + + PRCGV DI
Sbjct: 296 QFFGYLNSSTLHSNFTDAFTENLQSAIIEFQTNFNLNTTGQLDQDIYKIISKPRCGVPDI 475
Query: 128 INGTTTMNAGKDTETTSNSDSKLRFHTVSHFTVFPGQPRWPE-GKQELTYAFFPGNELTE 186
INGTTTM +T P +P W + L YAF P N +T+
Sbjct: 476 INGTTTMKNNFINKT------------------MPFKPWWRNVENRSLAYAFHPENNVTD 601
Query: 187 TVKSVFATAFARWSEVTTLKFTETTLYSGADIKIGFFNGDHGDGEPFDGSLGTLAHAF-- 244
VKS+F AF RWS T L F ET ++ +DI+I F DG GT+ ++
Sbjct: 602 NVKSLFQDAFNRWSNATELNFIETMSFNDSDIRIAFLT--------LDGKGGTVGGSYIN 757
Query: 245 -SPRNGRFHLDAAEDWVVSGDVSKSSLPTAVDLESVAVHEIGHLLGLGHSSEEEAIMFPT 303
S G +LDA E WV+ + VDLESV +H++GHLLGLGHSS EEAIM+P
Sbjct: 758 SSVNVGSVYLDADEQWVLPSENVVEE--DDVDLESVVMHQVGHLLGLGHSSVEEAIMYPI 931
Query: 304 ISSRMKKVVLADDDVRGIQYLYGTN 328
+ K ++ DD++ IQ +YG N
Sbjct: 932 VLQEKKIELVNVDDLQRIQEIYGVN 1006
>TC79334 MtN9
Length = 1241
Score = 166 bits (419), Expect = 2e-41
Identities = 117/321 (36%), Positives = 156/321 (48%), Gaps = 3/321 (0%)
Frame = +1
Query: 8 FPLTISFSLTLLIVSARLFPDVPSWIP-PGTLPVGPWDAFRNFTGCRQGENYNGLSNLKN 66
+ + SL +IV+ L +P P PG +Q E GLS +K
Sbjct: 1 YQFELLLSLLFIIVNTTLSGYIPQLSPSPG----------------KQTEEIQGLSKIKQ 132
Query: 67 YFQHFGYIPRSPKSNFSDDFDDDLQEAIKTYQKNFNLNVTGELDDMTLRQVMLPRCGVAD 126
+ HF Y+ F D D+ AIK YQ+ FNL VTG LD TL+Q+MLPRCGV D
Sbjct: 133 HLYHFKYLQGLYLVGFDDYLDNKTISAIKAYQQFFNLQVTGHLDTETLQQIMLPRCGVPD 312
Query: 127 IINGTTTMNAGKDTETTSNSDSKLRFHTVSHFTVFPGQPRWPEGKQELTYAFFPGNELTE 186
I N D F F G +P+G +ELTY F P ++++
Sbjct: 313 I-----------------NPDINPDF----GFARAQGNKWFPKGTKELTYGFLPESKISI 429
Query: 187 TVKSVFATAFARWSEVT-TLKFTETTLYSGADIKIGFFNGDHGDGEPFDGSLGTLAHAFS 245
+VF AF RWS+ T LKF+E T Y ADIKIGF+N + E D + +
Sbjct: 430 DKVNVFRNAFTRWSQTTRVLKFSEATSYDDADIKIGFYNISYNSKEVIDVVVSDF--FIN 603
Query: 246 PRNGRFHLDAAEDWVVSGDVSKSSLPTAVDLESVAVHEIGHLLGLGHSSEEEAIMFPTI- 304
R+ L+A++ W DLE+VA+H+IGHLLGL HSS+ E+IM+PTI
Sbjct: 604 LRSFTIRLEASKVW---------------DLETVAMHQIGHLLGLDHSSDVESIMYPTIV 738
Query: 305 SSRMKKVVLADDDVRGIQYLY 325
KKV + D + IQ LY
Sbjct: 739 PLHQKKVQITVSDNQAIQQLY 801
>TC86917 similar to GP|3776080|emb|CAA77093.1 MtN9 {Medicago truncatula},
partial (59%)
Length = 1294
Score = 147 bits (371), Expect = 6e-36
Identities = 108/323 (33%), Positives = 163/323 (50%), Gaps = 8/323 (2%)
Frame = +1
Query: 58 YNGLSNLKNYFQHFGYIPRSPKSNFSDDFDDDLQEAIKTYQKNFNLNVTGELDDMTLRQV 117
Y G+ +K Y FGY+ +S F++ D + A+KTYQ+ FN+ L ++ L+ +
Sbjct: 337 YKGVDQIKQYLSDFGYLEQS--GPFNNTLDQETVLALKTYQRYFNIQ-QDTLSEI-LQHI 504
Query: 118 MLPRCGVADIINGTTTMNAGKDTETTSNSDSKLRFHTVSHFTVFPGQPRWPEGKQELTYA 177
LPRCGV D I L+++ + + G +P+G + LTY
Sbjct: 505 ALPRCGVPDRI---------------------LKYNLTNDISFPKGNQWFPKGTKNLTYG 621
Query: 178 FFPGNELTETVKSVFATAFARWSEVT-TLKFTETTLYSGADIKIGFFN--GDHGDGEPFD 234
F P N++ + +VF TA +WS T L FTET Y A+IKIGF+N D G +
Sbjct: 622 FDPRNKIPLDMTNVFRTALTQWSNTTRVLNFTETKSYDDANIKIGFYNITDDDGINDVAV 801
Query: 235 GSLGTLAHAFSPRNGRFHLDAAEDWVVSGDVSKSSLPT---AVDLESVAVHEIGHLLGLG 291
G + + + ++G LDA + W +LPT DLE+ A+H+IGHLLGL
Sbjct: 802 GFTFIVLDSTNVKSGFITLDATKYW---------ALPTEHRGFDLETAAMHQIGHLLGLE 954
Query: 292 HSSEEEAIMFPTI-SSRMKKVVLADDDVRGIQYLYGTNPSFNGSTVISSP-ERNIGNGGC 349
HSS+ ++IM+PTI S K V + D D IQ LY ++ N ++ SS + G+
Sbjct: 955 HSSDNKSIMYPTILPSHQKNVQITDSDNLAIQKLYSSSTKANANSDDSSGCFKLFGSSSS 1134
Query: 350 SLTSLWSPWRLFSLLTFVLSHFL 372
L SL + +LL L++ L
Sbjct: 1135LLISLSIVFAFVALLN*HLNYIL 1203
>TC81657 weakly similar to PIR|T51957|T51957 metalloproteinase (EC 3.4.24.-)
[imported] - Arabidopsis thaliana, partial (24%)
Length = 537
Score = 144 bits (362), Expect = 7e-35
Identities = 72/108 (66%), Positives = 85/108 (78%)
Frame = +2
Query: 248 NGRFHLDAAEDWVVSGDVSKSSLPTAVDLESVAVHEIGHLLGLGHSSEEEAIMFPTISSR 307
+GR HLD AEDWVV+GDV++SSL AVDLESV VHEIGHLLGLGHSS EEAIM+PTISSR
Sbjct: 5 DGRLHLDKAEDWVVNGDVTESSLSNAVDLESVVVHEIGHLLGLGHSSIEEAIMYPTISSR 184
Query: 308 MKKVVLADDDVRGIQYLYGTNPSFNGSTVISSPERNIGNGGCSLTSLW 355
KKV L DD+ GIQ LYG+NP+F G+T +S +R+ GG SL+
Sbjct: 185 TKKVELESDDIEGIQMLYGSNPNFTGTTTATSRDRDSSFGGRHGVSLF 328
>TC86916 similar to GP|3776080|emb|CAA77093.1 MtN9 {Medicago truncatula},
partial (39%)
Length = 954
Score = 101 bits (251), Expect(2) = 3e-31
Identities = 61/174 (35%), Positives = 86/174 (49%), Gaps = 1/174 (0%)
Frame = +2
Query: 58 YNGLSNLKNYFQHFGYIPRSPKSNFSDDFDDDLQEAIKTYQKNFNLNVTGELDDMTLRQV 117
Y GL +K Y Q+FGY+ +S F++ D + A+KTYQ+ FN+ + L+ +
Sbjct: 260 YKGLDQIKQYLQNFGYLEQS--GPFNNTLDQETVLALKTYQRYFNIYAGQDSLRKILQHI 433
Query: 118 MLPRCGVADIINGTTTMNAGKDTETTSNSDSKLRFHTVSHFTVFPGQPRWPEGKQELTYA 177
LPRCGV D+ + + + + G +P+G + LTY
Sbjct: 434 ALPRCGVPDM---------------------NFTYDSTNDISYPKGNQWFPKGTKNLTYG 550
Query: 178 FFPGNELTETVKSVFATAFARWSEVT-TLKFTETTLYSGADIKIGFFNGDHGDG 230
F P NE+ V +VF A RWS+ T L FTETT Y ADIKI F N + DG
Sbjct: 551 FAPKNEIPLNVTNVFRKALTRWSQTTRVLNFTETTSYDDADIKIVFNNMTYDDG 712
Score = 52.0 bits (123), Expect(2) = 3e-31
Identities = 28/60 (46%), Positives = 40/60 (66%), Gaps = 1/60 (1%)
Frame = +1
Query: 253 LDAAEDWVVSGDVSKSSLPTAVDLESVAVHEIGHLLGLGHSSEEEAIMFPTI-SSRMKKV 311
LD + WV + + +DLE+ A+H+IGHLLGL HSS+ ++IM+PTI S+ KKV
Sbjct: 787 LDITKHWVFPTEDGE------LDLETAAMHQIGHLLGLEHSSDSKSIMYPTILPSQQKKV 948
>TC81934 weakly similar to PIR|E71433|E71433 probable metalloproteinase (EC
3.4.24.-) - Arabidopsis thaliana, partial (24%)
Length = 619
Score = 95.9 bits (237), Expect(2) = 3e-23
Identities = 56/161 (34%), Positives = 76/161 (46%), Gaps = 4/161 (2%)
Frame = +1
Query: 14 FSLTLLI---VSARLFPDVPSWIPPGTLPVGPWDAFRNFTGCRQGENYNGLSNLKNYFQH 70
F +TL + AR+ P+ + I W F F G + +G++ LK YF
Sbjct: 70 FFMTLFLRPCFPARIIPESVTVITTTQKHNATWHDFSRFLHAEPGSHVSGMAELKKYFNR 249
Query: 71 FGYI-PRSPKSNFSDDFDDDLQEAIKTYQKNFNLNVTGELDDMTLRQVMLPRCGVADIIN 129
FGY+ P +NF+D FD Q A+ YQKN L +TG+LD T+ ++ PRCGV+
Sbjct: 250 FGYLSPPLTTANFTDTFDSSFQSAVVLYQKNLGLPITGKLDSNTISTIVSPRCGVS---- 417
Query: 130 GTTTMNAGKDTETTSNSDSKLRFHTVSHFTVFPGQPRWPEG 170
DT T R HT HF F G+PRW G
Sbjct: 418 ---------DTAATH------RIHTTQHFAYFNGKPRWLRG 495
Score = 30.0 bits (66), Expect(2) = 3e-23
Identities = 15/33 (45%), Positives = 21/33 (63%), Gaps = 4/33 (12%)
Frame = +2
Query: 174 LTYAFFPGNELT----ETVKSVFATAFARWSEV 202
LTYAF P N + +++VF AFARW++V
Sbjct: 509 LTYAFSPYNMIDTLSLSEIQTVFERAFARWAKV 607
>TC82218 similar to GP|7159629|emb|CAB76364.1 matrix metalloproteinase
{Cucumis sativus}, partial (19%)
Length = 688
Score = 60.5 bits (145), Expect(2) = 1e-18
Identities = 26/44 (59%), Positives = 29/44 (65%)
Frame = +3
Query: 216 ADIKIGFFNGDHGDGEPFDGSLGTLAHAFSPRNGRFHLDAAEDW 259
ADIKIGF G+HGD PFDG LAH F P +GR H D E+W
Sbjct: 24 ADIKIGFHRGNHGDVYPFDGPGNVLAHTFPPEDGRLHFDGDENW 155
Score = 50.4 bits (119), Expect(2) = 1e-18
Identities = 28/59 (47%), Positives = 38/59 (63%), Gaps = 4/59 (6%)
Frame = +1
Query: 271 PTAV----DLESVAVHEIGHLLGLGHSSEEEAIMFPTISSRMKKVVLADDDVRGIQYLY 325
PTA DLE+VA+HE+GHLLGL HS+++ + M+P + + L DDV GI LY
Sbjct: 238 PTATTKQFDLETVALHEMGHLLGLAHSTDQNSAMYPYWAGVRRN--LNQDDVDGITALY 408
>BG581966 similar to GP|3776080|emb MtN9 {Medicago truncatula}, partial (53%)
Length = 683
Score = 89.7 bits (221), Expect = 2e-18
Identities = 59/147 (40%), Positives = 80/147 (54%), Gaps = 6/147 (4%)
Frame = +1
Query: 192 FATAFARWSEVT-TLKFTETTLYSGADIKIGFF---NGDHGDGEPFDGSLGTLAHAFSPR 247
F AF RWS+ T L F+ETT Y ADIKIGF+ N D D S + + +
Sbjct: 19 FRNAFTRWSQTTRVLNFSETTSYDDADIKIGFYHIYNNDIVDDVVVGDSFISRNLDSNVK 198
Query: 248 NGRFHLDAAEDWVV-SGDVSKSSLPTAVDLESVAVHEIGHLLGLGHSSEEEAIMFPTISS 306
+G L+ ++ WV + K DLE+V +H+IGHLLGL HSS++E+IM+P I
Sbjct: 199 SGMIRLERSKFWVSPTTTYFKKWELQEFDLETVVMHQIGHLLGLDHSSDKESIMYPLIDP 378
Query: 307 -RMKKVVLADDDVRGIQYLYGTNPSFN 332
+ KKV + D D + IQ LY N
Sbjct: 379 LQEKKVQITDSDNQAIQQLYTNTAKAN 459
>BF633849 weakly similar to PIR|E71433|E714 probable metalloproteinase (EC
3.4.24.-) - Arabidopsis thaliana, partial (5%)
Length = 544
Score = 79.0 bits (193), Expect = 3e-15
Identities = 40/138 (28%), Positives = 70/138 (49%), Gaps = 4/138 (2%)
Frame = +2
Query: 45 AFRNFTGCRQGENYNGLSNLKNYFQHFGYI----PRSPKSNFSDDFDDDLQEAIKTYQKN 100
+ +N G ++G+ G+ LKNY + FGY+ +P + +D FD++L+ A+K +Q
Sbjct: 143 SLQNLKGVQKGQTQKGIGELKNYLKRFGYLNVQHDATPSTTLNDHFDENLEFALKDFQTY 322
Query: 101 FNLNVTGELDDMTLRQVMLPRCGVADIINGTTTMNAGKDTETTSNSDSKLRFHTVSHFTV 160
+L+VTG +D T++ + LPRCGV D+ +S + S +
Sbjct: 323 HHLHVTGRVDTTTIKILSLPRCGVPDL---------------PKHSHKQNGLQMSSSYAF 457
Query: 161 FPGQPRWPEGKQELTYAF 178
F P+W + K+ L Y +
Sbjct: 458 FQDSPKWSDTKRNLKYMY 511
>AL379682 weakly similar to SP|P29136|MEP1_ Metalloendoproteinase 1 precursor
(EC 3.4.24.-) (SMEP1). [Soybean] {Glycine max}, partial
(15%)
Length = 386
Score = 73.6 bits (179), Expect = 1e-13
Identities = 37/75 (49%), Positives = 45/75 (59%)
Frame = +3
Query: 53 RQGENYNGLSNLKNYFQHFGYIPRSPKSNFSDDFDDDLQEAIKTYQKNFNLNVTGELDDM 112
+Q E GLS +K + HF Y+ F D D+ AIK YQ+ FNL VTG LD
Sbjct: 81 KQTEEIQGLSKIKQHLYHFKYLQGLYLVGFDDYLDNKTISAIKAYQQFFNLQVTGHLDTE 260
Query: 113 TLRQVMLPRCGVADI 127
TL+Q+MLPRCGV DI
Sbjct: 261 TLQQIMLPRCGVPDI 305
>AL373486 weakly similar to GP|16901508|gb| matrix metalloproteinase MMP2
{Glycine max}, partial (7%)
Length = 409
Score = 67.4 bits (163), Expect = 8e-12
Identities = 43/116 (37%), Positives = 56/116 (48%)
Frame = +2
Query: 7 IFPLTISFSLTLLIVSARLFPDVPSWIPPGTLPVGPWDAFRNFTGCRQGENYNGLSNLKN 66
+ P+ I SL +IV L +P P +G +Q E GLS +K
Sbjct: 5 VIPI*IVLSLLFIIVKTTLSGYIPQLSPS----LG-----------KQTEEIQGLSKIKQ 139
Query: 67 YFQHFGYIPRSPKSNFSDDFDDDLQEAIKTYQKNFNLNVTGELDDMTLRQVMLPRC 122
+ HF Y+ F D D+ AIK YQ+ FNL VTG LD TL+Q+MLPRC
Sbjct: 140 HLYHFKYLQGLYLVGFDDYLDNKTISAIKGYQQFFNLQVTGHLDTETLQQIMLPRC 307
>TC76470 similar to PIR|D84651|D84651 hypothetical protein At2g25680
[imported] - Arabidopsis thaliana, partial (53%)
Length = 1344
Score = 29.3 bits (64), Expect = 2.6
Identities = 13/29 (44%), Positives = 17/29 (57%)
Frame = -3
Query: 341 ERNIGNGGCSLTSLWSPWRLFSLLTFVLS 369
E NI NG C + L P+ FSL+ F+ S
Sbjct: 586 EFNIDNGSCGIAPLIKPFFQFSLVNFITS 500
>TC78730 similar to GP|13876520|gb|AAK43496.1 hypothetical protein {Oryza
sativa (japonica cultivar-group)}, partial (64%)
Length = 1068
Score = 28.9 bits (63), Expect = 3.3
Identities = 13/31 (41%), Positives = 17/31 (53%)
Frame = +1
Query: 227 HGDGEPFDGSLGTLAHAFSPRNGRFHLDAAE 257
HG G P S L+H +P + RF DAA+
Sbjct: 508 HGSGGPAGSSSSALSHPSAPFDARFSEDAAQ 600
>TC92483 similar to GP|21592537|gb|AAM64486.1 unknown {Arabidopsis
thaliana}, partial (8%)
Length = 662
Score = 28.1 bits (61), Expect = 5.7
Identities = 19/61 (31%), Positives = 34/61 (55%)
Frame = +1
Query: 89 DLQEAIKTYQKNFNLNVTGELDDMTLRQVMLPRCGVADIINGTTTMNAGKDTETTSNSDS 148
+L ++ K+ +K FN +T ++ T+ + + IN TTT + K T+TTSN +S
Sbjct: 199 NL*QSAKS*KKKFNQEITTKITFTTI----ITTTSNQNFIN-TTTFSKAKVTKTTSNKES 363
Query: 149 K 149
+
Sbjct: 364 Q 366
>AW683199 similar to PIR|B85043|B85 probable GH3-like protein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 522
Score = 28.1 bits (61), Expect = 5.7
Identities = 22/70 (31%), Positives = 34/70 (48%), Gaps = 4/70 (5%)
Frame = +1
Query: 308 MKKVVLADDDVRGIQYLYGTN--PSFNGSTVISSPERNIGNGGCSLTSLWSP--WRLFSL 363
MK +++ G Q L+ F T++ + RN G ++ S W+ W LFSL
Sbjct: 13 MKTLLVGSKMCLGTQVLFKLRYYAIF*NKTMVWNISRN--G*GITIYSKWNHVHWNLFSL 186
Query: 364 LTFVLSHFLL 373
L F+L H L+
Sbjct: 187 LWFLLLHMLI 216
>TC77078 similar to GP|21537078|gb|AAM61419.1 putaive DNA-binding protein
{Arabidopsis thaliana}, partial (51%)
Length = 1346
Score = 27.7 bits (60), Expect = 7.4
Identities = 16/56 (28%), Positives = 23/56 (40%)
Frame = +2
Query: 213 YSGADIKIGFFNGDHGDGEPFDGSLGTLAHAFSPRNGRFHLDAAEDWVVSGDVSKS 268
YS + F + GDG DG +G + + P H + +SG VS S
Sbjct: 587 YSISTTTSSFMSTITGDGSVSDGKIGPIISSGKPPLASSHRKRCHEATISGKVSSS 754
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.137 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,748,823
Number of Sequences: 36976
Number of extensions: 161952
Number of successful extensions: 846
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 823
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 827
length of query: 373
length of database: 9,014,727
effective HSP length: 98
effective length of query: 275
effective length of database: 5,391,079
effective search space: 1482546725
effective search space used: 1482546725
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146588.9


