

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146588.4 - phase: 0
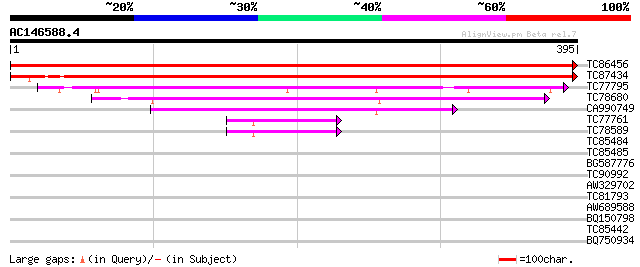
(395 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86456 similar to SP|P52902|ODPA_PEA Pyruvate dehydrogenase E1 ... 800 0.0
TC87434 similar to SP|P52902|ODPA_PEA Pyruvate dehydrogenase E1 ... 655 0.0
TC77795 similar to GP|15450707|gb|AAK96625.1 At1g01090/T25K16_8 ... 246 1e-65
TC78680 similar to GP|9454571|gb|AAF87894.1| branched-chain alph... 151 5e-37
CA990749 similar to GP|9955517|emb branched-chain alpha keto-aci... 113 1e-25
TC77761 1-deoxy-D-xylulose 5-phosphate synthase 1 [Medicago trun... 42 4e-04
TC78589 1-deoxy-D-xylulose 5-phosphate synthase 2 [Medicago trun... 42 5e-04
TC85484 similar to PIR|S58083|S58083 transketolase (EC 2.2.1.1) ... 33 0.15
TC85485 similar to GP|22136900|gb|AAM91794.1 putative transketol... 32 0.33
BG587776 weakly similar to PIR|F86242|F86 unknown protein 98896... 30 2.1
TC90992 similar to PIR|F86242|F86242 unknown protein 98896-9585... 30 2.1
AW329702 29 2.8
TC81793 weakly similar to PIR|D86165|D86165 protein F15K9.3 [imp... 29 3.6
AW689588 similar to GP|9757805|dbj CCR4-associated factor-like p... 28 4.7
BQ150798 28 6.2
TC85442 similar to GP|13543783|gb|AAH06040.1 Unknown (protein fo... 28 6.2
BQ750934 similar to PIR|T19673|T19 hypothetical protein C33B4.3 ... 28 8.0
>TC86456 similar to SP|P52902|ODPA_PEA Pyruvate dehydrogenase E1 component
alpha subunit mitochondrial precursor (EC 1.2.4.1)
(PDHE1-A)., partial (92%)
Length = 1781
Score = 800 bits (2065), Expect = 0.0
Identities = 395/395 (100%), Positives = 395/395 (100%)
Frame = +2
Query: 1 MALSRFSSSQFGSTLLKPYFLSSAIRHRSISSSSTETLTVETSIPFTSHNCEPPSTTVQT 60
MALSRFSSSQFGSTLLKPYFLSSAIRHRSISSSSTETLTVETSIPFTSHNCEPPSTTVQT
Sbjct: 203 MALSRFSSSQFGSTLLKPYFLSSAIRHRSISSSSTETLTVETSIPFTSHNCEPPSTTVQT 382
Query: 61 TASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDCVIT 120
TASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDCVIT
Sbjct: 383 TASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDCVIT 562
Query: 121 AYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIPLG 180
AYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIPLG
Sbjct: 563 AYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIPLG 742
Query: 181 VGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEW 240
VGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEW
Sbjct: 743 VGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEW 922
Query: 241 RSAKSPAYYKRGDYAPGLKVDGMDVLAVKQACKFAKEHALKNGPLILEMDTYRYHGHSMS 300
RSAKSPAYYKRGDYAPGLKVDGMDVLAVKQACKFAKEHALKNGPLILEMDTYRYHGHSMS
Sbjct: 923 RSAKSPAYYKRGDYAPGLKVDGMDVLAVKQACKFAKEHALKNGPLILEMDTYRYHGHSMS 1102
Query: 301 DPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKE 360
DPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKE
Sbjct: 1103DPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKE 1282
Query: 361 SQMPDPSDLFTNVYVKGLGVEACGADRKEVKATLP 395
SQMPDPSDLFTNVYVKGLGVEACGADRKEVKATLP
Sbjct: 1283SQMPDPSDLFTNVYVKGLGVEACGADRKEVKATLP 1387
>TC87434 similar to SP|P52902|ODPA_PEA Pyruvate dehydrogenase E1 component
alpha subunit mitochondrial precursor (EC 1.2.4.1)
(PDHE1-A)., complete
Length = 1829
Score = 655 bits (1691), Expect = 0.0
Identities = 325/398 (81%), Positives = 358/398 (89%), Gaps = 3/398 (0%)
Frame = +2
Query: 1 MALSRFSSSQFG---STLLKPYFLSSAIRHRSISSSSTETLTVETSIPFTSHNCEPPSTT 57
MALSR SSS S L+KP + +RSISS +T L++ETSIPFT+HNC PPSTT
Sbjct: 230 MALSRLSSSSTSTIKSNLIKPISTIFTL-NRSISSDTT--LSIETSIPFTAHNCTPPSTT 400
Query: 58 VQTTASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDC 117
V TT +EL++FF+ M LMRRMEIAADSLYK+KLIRGFCHLYDGQEAVAVGMEA+ +KDC
Sbjct: 401 VTTTPNELLNFFHTMSLMRRMEIAADSLYKSKLIRGFCHLYDGQEAVAVGMEASTTKKDC 580
Query: 118 VITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQI 177
+ITAYRDHCTFL RGGTL+EV+SELMGR DGCSKGKGGSMHFY+K+ GF+GGHGIVGAQ+
Sbjct: 581 IITAYRDHCTFLGRGGTLLEVYSELMGRVDGCSKGKGGSMHFYKKDSGFFGGHGIVGAQV 760
Query: 178 PLGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGT 237
PLG GLAFGQKY K+ +VTF LYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGT
Sbjct: 761 PLGCGLAFGQKYLKNESVTFALYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGT 940
Query: 238 AEWRSAKSPAYYKRGDYAPGLKVDGMDVLAVKQACKFAKEHALKNGPLILEMDTYRYHGH 297
AEWR+AKSPAYYKRGDY PGLKVDGMDVLAVKQA KFAKEHAL+NGP+ILEMDTYRYHGH
Sbjct: 941 AEWRAAKSPAYYKRGDYVPGLKVDGMDVLAVKQAVKFAKEHALQNGPIILEMDTYRYHGH 1120
Query: 298 SMSDPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAK 357
SMSDPGSTYRTRDEISGVRQERDPIERV+KL+L+HDI+TEKELKD EKE RK+VDEAIAK
Sbjct: 1121SMSDPGSTYRTRDEISGVRQERDPIERVKKLLLSHDIATEKELKDTEKEVRKEVDEAIAK 1300
Query: 358 AKESQMPDPSDLFTNVYVKGLGVEACGADRKEVKATLP 395
AKES +P PSDLFTNVYVKG GVEA GADRKEV+ATLP
Sbjct: 1301AKESPVPKPSDLFTNVYVKGYGVEAFGADRKEVRATLP 1414
>TC77795 similar to GP|15450707|gb|AAK96625.1 At1g01090/T25K16_8
{Arabidopsis thaliana}, partial (82%)
Length = 1690
Score = 246 bits (628), Expect = 1e-65
Identities = 149/398 (37%), Positives = 215/398 (53%), Gaps = 28/398 (7%)
Frame = +2
Query: 20 FLSSAIRHRSISSS---STETLTVETSIPFTSHNCEPPSTTV---QT-----TASELMSF 68
F S +H + SSS ST+ L + + PP V QT T E +
Sbjct: 269 FSSDLFKHNASSSSFLGSTQKL-----LRLNARRSPPPVAAVLLQQTSNLLITKEEGLVL 433
Query: 69 FNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDCVITAYRDHCTF 128
+ DMVL R E +Y + GF HLY+GQEAV+ G + ++DC+++ YRDH
Sbjct: 434 YEDMVLGRSFEDKCAEMYYRGKMFGFVHLYNGQEAVSTGFIKYLRKEDCIVSTYRDHVHA 613
Query: 129 LCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIPLGVGLAFGQK 188
L +G EV SEL G+ GC +G+GGSMH + KE GG +G IP+ G AF K
Sbjct: 614 LSKGVPSREVMSELFGKATGCCRGQGGSMHMFSKEHNVLGGFAFIGEGIPVATGAAFSMK 793
Query: 189 YNKD-------PNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWR 241
Y ++ NVT +GDG N GQ FE LN+AALW LP + V ENN + +G + R
Sbjct: 794 YKREVLNQADSDNVTLAFFGDGTCNNGQFFECLNMAALWKLPIVFVVENNLWAIGMSHLR 973
Query: 242 SAKSPAYYKRGDY--APGLKVDGMDVLAVKQACKFAKEHALK-NGPLILEMDTYRYHGHS 298
+ P +K+G PG+ VDGMDVL V++ K A + A + GP ++E +TYR+ GHS
Sbjct: 974 ATSDPEIWKKGPAFGMPGVHVDGMDVLKVREVAKEAIDRARRGEGPSLVECETYRFRGHS 1153
Query: 299 MSDPGSTYRTRDEISGVRQE-----RDPIERVRKLVLAHDISTEKELKDIEKEARKQVDE 353
++DP DE+ ++ RDPI ++K + + ++TE+ELK I+K+ + ++E
Sbjct: 1154LADP-------DELRNPAEKQHYAGRDPITALKKYLFENKLATEQELKAIDKKIDEVLEE 1312
Query: 354 AIAKAKESQMPDPSDLFTNVYV--KGLGVEACGADRKE 389
A+ A++S P S L NV+ KG G+ G R E
Sbjct: 1313AVDFAEKSPAPARSQLLENVFADPKGFGIGPDGRYRCE 1426
>TC78680 similar to GP|9454571|gb|AAF87894.1| branched-chain alpha keto-acid
dehydrogenase E1 - alpha subunit {Arabidopsis thaliana},
partial (76%)
Length = 1520
Score = 151 bits (381), Expect = 5e-37
Identities = 96/325 (29%), Positives = 161/325 (49%), Gaps = 6/325 (1%)
Frame = +3
Query: 58 VQTTASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLY---DGQEAVAVGMEAAINR 114
VQ + + +++MV ++ M DS++ +G Y G+EAV + AA++
Sbjct: 354 VQVSKEMAVRMYSEMVTLQTM----DSIFYEVQRQGRISFYLTSMGEEAVNIASAAALSS 521
Query: 115 KDCVITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVG 174
D ++ YR+ L RG TL + +L G + KG+ +H+ ++ +
Sbjct: 522 DDIILPQYREPGVLLWRGFTLQQFAHQLFGNTNDFGKGRQMPIHYGSNNHNYFTVSSPIA 701
Query: 175 AQIPLGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNHYG 234
Q+P VG A+ K + T GDG ++G +N AA+ + P I +C NN +
Sbjct: 702 TQLPQAVGAAYSLKMDGKSACAVTFCGDGGTSEGDFHAGMNFAAVMEAPVIFICRNNGWA 881
Query: 235 MGTAEWRSAKSPAYYKRGDYAP--GLKVDGMDVLAVKQACKFAKEHALK-NGPLILEMDT 291
+ T +S +G ++VDG D LAV A A+E A+K P+++E +
Sbjct: 882 ISTPVEEQFRSDGIVVKGQAYGIWSIRVDGNDALAVYSAVHTAREIAIKEQRPVLIEALS 1061
Query: 292 YRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQV 351
YR HS SD + YR DEI + ER+P+ R +K V + ++K+ ++ RKQ+
Sbjct: 1062YRVGHHSTSDDSTKYRPIDEIEYWKMERNPVNRFKKWVERNGWWSDKDELELRSSVRKQL 1241
Query: 352 DEAIAKAKESQMPDPSDLFTNVYVK 376
+AI A+++Q P D+FT+VY K
Sbjct: 1242MQAIQVAEKAQKPPLEDMFTDVYDK 1316
>CA990749 similar to GP|9955517|emb branched-chain alpha keto-acid
dehydrogenase E1 alpha subunit-like protein {Arabidopsis
thaliana}, partial (53%)
Length = 799
Score = 113 bits (283), Expect = 1e-25
Identities = 66/217 (30%), Positives = 106/217 (48%), Gaps = 3/217 (1%)
Frame = +2
Query: 99 DGQEAVAVGMEAAINRKDCVITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMH 158
+G+EA+ + AA++ D + YR+ L RG TL E ++ K KG+ H
Sbjct: 17 NGEEAINIASAAALSMNDVIFPQYREQGVLLWRGFTLQEFANQCFSNKFDNGKGRQMPAH 196
Query: 159 FYRKEGGFYGGHGIVGAQIPLGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAA 218
+ + + V QIP VG A+ K +K T +GDG +++G LN AA
Sbjct: 197 YGSNKHNYMNVASTVATQIPHAVGAAYSLKMDKKDACAVTYFGDGGSSEGDFHAGLNFAA 376
Query: 219 LWDLPAILVCENNHYGMGTAEWRSAKSPAYYKRGDY--APGLKVDGMDVLAVKQACKFAK 276
+ + P I +C NN + + T +S +G ++VDG D LA+ A + A+
Sbjct: 377 VMEAPVIFICRNNGWAISTPTSDQFRSDGIVVKGQAYGVRSIRVDGNDALAIYSAVQAAR 556
Query: 277 EHAL-KNGPLILEMDTYRYHGHSMSDPGSTYRTRDEI 312
+ A+ + P+++E TYR HS SD + YR +EI
Sbjct: 557 QMAVSEERPILIEALTYRVGHHSTSDDSTKYRPANEI 667
>TC77761 1-deoxy-D-xylulose 5-phosphate synthase 1 [Medicago truncatula]
Length = 2595
Score = 42.0 bits (97), Expect = 4e-04
Identities = 27/92 (29%), Positives = 38/92 (40%), Gaps = 12/92 (13%)
Frame = +3
Query: 152 GKGGSMHFYRKEGGFYG------------GHGIVGAQIPLGVGLAFGQKYNKDPNVTFTL 199
G+ MH R+ G G G G I G+G+A G+ N +
Sbjct: 582 GRRDKMHTMRQTNGLSGFTKRSESEYDSFGTGHSSTTISAGLGMAVGRDLKGRKNDVVAV 761
Query: 200 YGDGAANQGQLFEALNIAALWDLPAILVCENN 231
GDGA GQ +EA+N A D I++ +N
Sbjct: 762 IGDGAMTAGQAYEAMNNAGYLDSDMIVILNDN 857
>TC78589 1-deoxy-D-xylulose 5-phosphate synthase 2 [Medicago truncatula]
Length = 2760
Score = 41.6 bits (96), Expect = 5e-04
Identities = 27/92 (29%), Positives = 38/92 (40%), Gaps = 12/92 (13%)
Frame = +3
Query: 152 GKGGSMHFYRKEGGFYG------------GHGIVGAQIPLGVGLAFGQKYNKDPNVTFTL 199
G+ MH RK G G G G I G+G+A + N ++
Sbjct: 630 GRRSRMHTIRKTSGLAGFPKRDESVHDAFGVGHSSTSISAGLGMAIARDLLGKKNSVISV 809
Query: 200 YGDGAANQGQLFEALNIAALWDLPAILVCENN 231
GDGA GQ +EA+N A D I++ +N
Sbjct: 810 IGDGAMTAGQAYEAMNNAGFIDSNLIVILNDN 905
>TC85484 similar to PIR|S58083|S58083 transketolase (EC 2.2.1.1) precursor -
potato (fragment), partial (95%)
Length = 2685
Score = 33.5 bits (75), Expect = 0.15
Identities = 37/161 (22%), Positives = 69/161 (41%), Gaps = 14/161 (8%)
Frame = +1
Query: 171 GIVGAQIPLGVGLAFGQK-----YNKDPN-----VTFTLYGDGAANQGQLFEALNIAALW 220
G +G I VGLA +K +NK N T+ + GDG +G EA ++A W
Sbjct: 652 GPLGQGIANAVGLALAEKHLAARFNKPDNEIIDHYTYCILGDGCQMEGIANEACSLAGHW 831
Query: 221 DLPAILVCENNHYGMGTAEWRSAKSPAYYKR----GDYAPGLKVDGMDVLAVKQACKFAK 276
L ++ ++++ + A + + KR G + +K ++ A K AK
Sbjct: 832 GLGKLIAFYDDNHISIDGDTEIAFTESVEKRFEGLGWHVIWVKNGNTGFDEIRAAIKEAK 1011
Query: 277 EHALKNGPLILEMDTYRYHGHSMSDPGSTYRTRDEISGVRQ 317
A+K+ P +++ T G+ + ++Y G ++
Sbjct: 1012--AVKDRPTLIKFTT--TIGYGSPNKSNSYSVHGSALGAKE 1122
>TC85485 similar to GP|22136900|gb|AAM91794.1 putative transketolase
{Arabidopsis thaliana}, partial (90%)
Length = 2728
Score = 32.3 bits (72), Expect = 0.33
Identities = 23/73 (31%), Positives = 36/73 (48%), Gaps = 11/73 (15%)
Frame = +2
Query: 171 GIVGAQIPLGVGLAFGQK-----YNKDPN-----VTFTLYGDGAANQGQLFEALNIAALW 220
G +G I GVGLA +K +NK + T+ + GDG +G EA ++A W
Sbjct: 746 GPLGQGIANGVGLALAEKHLAARFNKPDSEIVDHYTYVILGDGCQMEGISNEACSLAGHW 925
Query: 221 DLPAILV-CENNH 232
L ++ ++NH
Sbjct: 926 GLGKLIAFYDDNH 964
>BG587776 weakly similar to PIR|F86242|F86 unknown protein 98896-95855
[imported] - Arabidopsis thaliana, partial (23%)
Length = 608
Score = 29.6 bits (65), Expect = 2.1
Identities = 20/72 (27%), Positives = 32/72 (43%)
Frame = +2
Query: 292 YRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQV 351
YR S +++ G +++ IE+ RK E ELK I +E K+V
Sbjct: 146 YRRQRSRSSSLSPAHKSSSSSLGSVEQKTAIEKQRKEEEKKRRQQEAELKLIAEETAKRV 325
Query: 352 DEAIAKAKESQM 363
+EAI K + +
Sbjct: 326 EEAIRKRVDESL 361
>TC90992 similar to PIR|F86242|F86242 unknown protein 98896-95855
[imported] - Arabidopsis thaliana, partial (14%)
Length = 763
Score = 29.6 bits (65), Expect = 2.1
Identities = 20/72 (27%), Positives = 32/72 (43%)
Frame = +1
Query: 292 YRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQV 351
YR S +++ G +++ IE+ RK E ELK I +E K+V
Sbjct: 457 YRRQRSRSSSLSPAHKSSSSSLGSVEQKTAIEKQRKEEEKKRRQQEAELKLIAEETAKRV 636
Query: 352 DEAIAKAKESQM 363
+EAI K + +
Sbjct: 637 EEAIRKRVDESL 672
>AW329702
Length = 563
Score = 29.3 bits (64), Expect = 2.8
Identities = 23/77 (29%), Positives = 42/77 (53%), Gaps = 2/77 (2%)
Frame = -3
Query: 19 YFLSSAIRHRSISSSSTETLTVETSIPFTSHNCEPPSTTVQTT--ASELMSFFNDMVLMR 76
+F+ S+ + +SSS ETLT+ +S+P T + STT Q + S S+ D+V++
Sbjct: 519 WFIVSSPKRLMVSSSPLETLTLLSSLPETITSSS--STT*QRSIFLSSGSSYDRDIVVLS 346
Query: 77 RMEIAADSLYKAKLIRG 93
M+ + + + L+ G
Sbjct: 345 AMKPSPKTTITSTLLPG 295
>TC81793 weakly similar to PIR|D86165|D86165 protein F15K9.3 [imported] -
Arabidopsis thaliana, partial (16%)
Length = 752
Score = 28.9 bits (63), Expect = 3.6
Identities = 16/60 (26%), Positives = 35/60 (57%), Gaps = 3/60 (5%)
Frame = +3
Query: 307 RTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEK---EARKQVDEAIAKAKESQM 363
+ + E+ +RQE++ +ER++K + +T K+L ++E +A QV+ A ++ +M
Sbjct: 243 KDKAELKTLRQEKEEVERLKKEKQCLEENTMKKLSEMENALGKAGGQVERANTAVRKLEM 422
>AW689588 similar to GP|9757805|dbj CCR4-associated factor-like protein
{Arabidopsis thaliana}, partial (46%)
Length = 527
Score = 28.5 bits (62), Expect = 4.7
Identities = 12/31 (38%), Positives = 19/31 (60%)
Frame = -1
Query: 31 SSSSTETLTVETSIPFTSHNCEPPSTTVQTT 61
SSS+ +T + IP++S N P S+ +Q T
Sbjct: 464 SSSTQQTSLTQPQIPYSSQNQSPDSSAIQLT 372
>BQ150798
Length = 920
Score = 28.1 bits (61), Expect = 6.2
Identities = 24/107 (22%), Positives = 47/107 (43%), Gaps = 4/107 (3%)
Frame = +2
Query: 223 PAILVCENNHYGMGTAEWRSAKSPAYYKRGDYAPGLKVDGMDVLAVKQACKFAKEHALKN 282
PA +VC + + R A S + A L+ G+D + ++ + ++ + H
Sbjct: 545 PAFVVCSSGGKVLARESTRHAVSVTIRQADAIADRLEEFGIDAVTIESSAQYRR*H---* 715
Query: 283 GPLILEMDTYRYHG---HSMSDPGSTY-RTRDEISGVRQERDPIERV 325
P +L+ + R +G H+++ P S DE+ V R P + +
Sbjct: 716 RPALLDTPSLRAYGAECHAVASPSSRP*VVLDEVEQVSYVRAPYQHM 856
>TC85442 similar to GP|13543783|gb|AAH06040.1 Unknown (protein for MGC:7642)
{Mus musculus}, partial (62%)
Length = 585
Score = 28.1 bits (61), Expect = 6.2
Identities = 16/31 (51%), Positives = 18/31 (57%)
Frame = -3
Query: 4 SRFSSSQFGSTLLKPYFLSSAIRHRSISSSS 34
S FSSS FGS+ P F SS+ S SSS
Sbjct: 196 SFFSSSFFGSSFFSPSFFSSSFFASSFFSSS 104
>BQ750934 similar to PIR|T19673|T19 hypothetical protein C33B4.3 -
Caenorhabditis elegans, partial (1%)
Length = 627
Score = 27.7 bits (60), Expect = 8.0
Identities = 24/75 (32%), Positives = 33/75 (44%), Gaps = 9/75 (12%)
Frame = +2
Query: 2 ALSRFSSSQFGSTLLKPYFLSSAIRHRSISSSST---ETLTVET------SIPFTSHNCE 52
+LS S++ T P F + A +SS+S+ ETL T SIP +S
Sbjct: 299 SLSTLSATNSSVTSAAPSFTAEAETTSPVSSASSTSLETLDATTTAVPSSSIPESSSTSL 478
Query: 53 PPSTTVQTTASELMS 67
PP+ Q T S S
Sbjct: 479 PPTLVPQNTTSPASS 523
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.135 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,649,675
Number of Sequences: 36976
Number of extensions: 162740
Number of successful extensions: 854
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 832
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 847
length of query: 395
length of database: 9,014,727
effective HSP length: 98
effective length of query: 297
effective length of database: 5,391,079
effective search space: 1601150463
effective search space used: 1601150463
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146588.4


