

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146587.10 + phase: 0 /pseudo
(1157 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
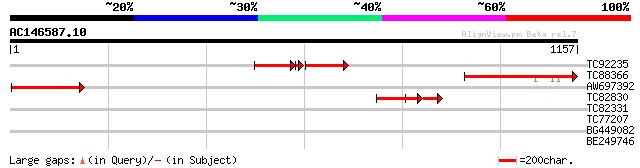
Score E
Sequences producing significant alignments: (bits) Value
TC92235 weakly similar to GP|15029315|gb|AAK81821.1 CYFIP2 {Mus ... 177 4e-93
TC88366 similar to GP|18419611|gb|AAL69375.1 unknown {Narcissus ... 315 8e-86
AW697392 295 5e-80
TC82830 186 4e-47
TC82331 weakly similar to EGAD|120604|128852 hypothetical protei... 31 3.2
TC77207 similar to GP|10334495|emb|CAC10209. putative extracellu... 31 3.2
BG449082 weakly similar to GP|7290650|gb| CG4020 gene product {D... 30 5.4
BE249746 weakly similar to PIR|T02553|T025 cellulose synthase ho... 30 7.0
>TC92235 weakly similar to GP|15029315|gb|AAK81821.1 CYFIP2 {Mus musculus},
partial (2%)
Length = 628
Score = 177 bits (450), Expect(3) = 4e-93
Identities = 86/88 (97%), Positives = 87/88 (98%)
Frame = +3
Query: 603 AVTVATLTDLGFLWFREFYLESSRVIQFPIECSLPWMLVDCVLESPNSGLLESVLMPFDI 662
+VTVATLTDLGFLWFREFYLESSRVIQFPIECSLPWMLVDCVLESPNSGLLESVLMPFDI
Sbjct: 351 SVTVATLTDLGFLWFREFYLESSRVIQFPIECSLPWMLVDCVLESPNSGLLESVLMPFDI 530
Query: 663 YNDSAQQALVLLKQRFLYDEIEAEVDHC 690
YNDSA QALVLLKQRFLYDEIEAEVDHC
Sbjct: 531 YNDSAXQALVLLKQRFLYDEIEAEVDHC 614
Score = 169 bits (429), Expect(3) = 4e-93
Identities = 83/84 (98%), Positives = 84/84 (99%)
Frame = +1
Query: 499 TFRKKKDLSRILSDMRTLSADWMANTNKSESELQSSQHGGEESKANIFYPRAVAPTAAQV 558
+FRKKKDLSRILSDMRTLSADWMANTNKSESELQSSQHGGEESKANIFYPRAVAPTAAQV
Sbjct: 7 SFRKKKDLSRILSDMRTLSADWMANTNKSESELQSSQHGGEESKANIFYPRAVAPTAAQV 186
Query: 559 HCLQFLIYEVVSGGNLRRPGGLFG 582
HCLQFLIYEVVSGGNLRRPGGLFG
Sbjct: 187 HCLQFLIYEVVSGGNLRRPGGLFG 258
Score = 35.4 bits (80), Expect(3) = 4e-93
Identities = 16/16 (100%), Positives = 16/16 (100%)
Frame = +2
Query: 584 SGSEVPVNDLKQLETF 599
SGSEVPVNDLKQLETF
Sbjct: 263 SGSEVPVNDLKQLETF 310
>TC88366 similar to GP|18419611|gb|AAL69375.1 unknown {Narcissus
pseudonarcissus}, partial (78%)
Length = 1001
Score = 315 bits (806), Expect = 8e-86
Identities = 170/255 (66%), Positives = 180/255 (69%), Gaps = 25/255 (9%)
Frame = +1
Query: 928 NEFHANCSLAGLASWSRWTNITFSRWWR*PCCQSL*IYSCCNGFIPRLPITCFFPHHVKT 987
NEFHANCSLAGLASWSRWTNITFSRWWR*PCCQSL*IYSCCNGFIPRLPITCFFPHHVKT
Sbjct: 1 NEFHANCSLAGLASWSRWTNITFSRWWR*PCCQSL*IYSCCNGFIPRLPITCFFPHHVKT 180
Query: 988 SRSCRSFI*G*LEYWKCSGVCACFHKCCIR*IL**MECCAQDRLH*HYNFKRFLPDIQWS 1047
SRSCRSFI*G*LEYWKCSGVCACFHKCCIR*IL**MECCAQDRLH*HYNFKRFLPDIQWS
Sbjct: 181 SRSCRSFI*G*LEYWKCSGVCACFHKCCIR*IL**MECCAQDRLH*HYNFKRFLPDIQWS 360
Query: 1048 PNCT*GYLEESAQVQSSSPERLG-------DSVAWGGCTI--IYLLGQQLHFELFDFSYQ 1098
PN G + S S R + AW T + LL +
Sbjct: 361 PNWILGRVCPSTIK*S*KTRRFSCLGGLHHNLFAWATATF*AL*LLVSDPQYRRSGSCIC 540
Query: 1099 ILNIAEVE-----AASVVQTQKNSH-----------FAVQP*RKDSMRY*AKWCSITSYK 1142
N E+ S+++ + S + P*RKDSMRY*AKWCSITSYK
Sbjct: 541 RANTKELSLCCSGMGSLIRGNEKSKEIK*PCILHA*SPLPP*RKDSMRY*AKWCSITSYK 720
Query: 1143 I*KHSFSI*NSATER 1157
I*KHSFSI*NSATER
Sbjct: 721 I*KHSFSI*NSATER 765
>AW697392
Length = 454
Score = 295 bits (756), Expect = 5e-80
Identities = 150/150 (100%), Positives = 150/150 (100%)
Frame = +3
Query: 4 PVEEAIAALSTFSLEDEQPEVQGPGVWVSTERGATESPIEYCDVAAYRLSLSEDTKALNQ 63
PVEEAIAALSTFSLEDEQPEVQGPGVWVSTERGATESPIEYCDVAAYRLSLSEDTKALNQ
Sbjct: 3 PVEEAIAALSTFSLEDEQPEVQGPGVWVSTERGATESPIEYCDVAAYRLSLSEDTKALNQ 182
Query: 64 LSSLTQEGKEMASVLYTYRSCVKALPQLPDSMKQSQADLYLETYQVLDLEMSRLREIQRW 123
LSSLTQEGKEMASVLYTYRSCVKALPQLPDSMKQSQADLYLETYQVLDLEMSRLREIQRW
Sbjct: 183 LSSLTQEGKEMASVLYTYRSCVKALPQLPDSMKQSQADLYLETYQVLDLEMSRLREIQRW 362
Query: 124 QASASSKLATDMQRFSRPERRINGPTISHL 153
QASASSKLATDMQRFSRPERRINGPTISHL
Sbjct: 363 QASASSKLATDMQRFSRPERRINGPTISHL 452
>TC82830
Length = 669
Score = 186 bits (472), Expect = 4e-47
Identities = 93/94 (98%), Positives = 94/94 (99%)
Frame = +1
Query: 749 EKLLDVLKHSHELLSIDLSVDSFSLMLNEMQENISLVSFSSRLASQIWSEMQSDFLPNFI 808
+KLLDVLKHSHELLSIDLSVDSFSLMLNEMQENISLVSFSSRLASQIWSEMQSDFLPNFI
Sbjct: 148 QKLLDVLKHSHELLSIDLSVDSFSLMLNEMQENISLVSFSSRLASQIWSEMQSDFLPNFI 327
Query: 809 LCNTTQRFIRSSKTVPVQKPSIPSAKPSFYCGTQ 842
LCNTTQRFIRSSKTVPVQKPSIPSAKPSFYCGTQ
Sbjct: 328 LCNTTQRFIRSSKTVPVQKPSIPSAKPSFYCGTQ 429
Score = 86.7 bits (213), Expect = 5e-17
Identities = 49/75 (65%), Positives = 54/75 (71%)
Frame = +3
Query: 808 ILCNTTQRFIRSSKTVPVQKPSIPSAKPSFYCGTQPS*SYIKQDYFA*TNDNRITRIHAK 867
IL NT+ F RS+ +P+ + S PS*SYIKQDYFA*TNDNRITRIHAK
Sbjct: 474 ILWNTSYVFYRSTFGIPLFTLAY*S----------PS*SYIKQDYFA*TNDNRITRIHAK 623
Query: 868 INRPSSF*WRRDRLC 882
INRPSSF*WRRDRLC
Sbjct: 624 INRPSSF*WRRDRLC 668
>TC82331 weakly similar to EGAD|120604|128852 hypothetical protein M01F1.5
{Caenorhabditis elegans}, partial (10%)
Length = 848
Score = 30.8 bits (68), Expect = 3.2
Identities = 27/111 (24%), Positives = 49/111 (43%), Gaps = 19/111 (17%)
Frame = -2
Query: 68 TQEGKEMASVLYTYRSCVKALPQLPDSMKQSQADLY-LETYQVLDLEMSRLREIQRWQAS 126
+Q G + + R KA+P+ P +M+Q +D + L T +++ LEM + I W +
Sbjct: 844 SQRGDSGTTRMPRPRGMAKAMPR-PMTMRQEASDSWMLRTPKLMRLEMRMPKVIMSWYEA 668
Query: 127 ASS-----------KLATDMQRFSRPERRINGPTI-------SHLWSMLKL 159
++ + T RF P +N P I + +W+ML +
Sbjct: 667 TTAPRISLGEHSDWNMGTPTDRFPTPRPAMNLPIIMCTQVSMAVIWTMLPM 515
>TC77207 similar to GP|10334495|emb|CAC10209. putative extracellular dermal
glycoprotein {Cicer arietinum}, partial (32%)
Length = 1024
Score = 30.8 bits (68), Expect = 3.2
Identities = 18/52 (34%), Positives = 32/52 (60%)
Frame = +1
Query: 755 LKHSHELLSIDLSVDSFSLMLNEMQENISLVSFSSRLASQIWSEMQSDFLPN 806
+KHS SI+L++ SFS + + + I+L+SFS + Q +S ++ D + N
Sbjct: 4 IKHSSITNSIELTMASFSTLFSTLILTIALLSFSCS-SQQFFSPVEKDPITN 156
>BG449082 weakly similar to GP|7290650|gb| CG4020 gene product {Drosophila
melanogaster}, partial (25%)
Length = 623
Score = 30.0 bits (66), Expect = 5.4
Identities = 13/36 (36%), Positives = 23/36 (63%)
Frame = -2
Query: 777 EMQENISLVSFSSRLASQIWSEMQSDFLPNFILCNT 812
+ + +SL S S++ A++IW+ + SDF P +L T
Sbjct: 349 DSKPGLSLRSLSNKGAAKIWANLSSDFTPFLLLRRT 242
>BE249746 weakly similar to PIR|T02553|T025 cellulose synthase homolog
T26B15.10 - Arabidopsis thaliana, partial (7%)
Length = 383
Score = 29.6 bits (65), Expect = 7.0
Identities = 25/95 (26%), Positives = 41/95 (42%)
Frame = +1
Query: 671 LVLLKQRFLYDEIEAEVDHCFDIFVARLCETIFTYYKSWAASELLDPTFLFASENAEKYA 730
L+LL R +Y ++ F+A +CET FT + S P F+ + + +
Sbjct: 91 LLLLGYRVMYVN-----NYSIPWFIALICETWFTLSWIFTISTQWSPAFIKTNPDRLLQS 255
Query: 731 VQPMRLNMLLKMTRVKELEKLLDVLKHSHELLSID 765
VQ + L T ELE + + LL++D
Sbjct: 256 VQELPAVDLFVTTADDELEPAIITINTVLSLLALD 360
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.331 0.140 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 38,621,352
Number of Sequences: 36976
Number of extensions: 619638
Number of successful extensions: 3685
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 2751
Number of HSP's successfully gapped in prelim test: 127
Number of HSP's that attempted gapping in prelim test: 858
Number of HSP's gapped (non-prelim): 2983
length of query: 1157
length of database: 9,014,727
effective HSP length: 107
effective length of query: 1050
effective length of database: 5,058,295
effective search space: 5311209750
effective search space used: 5311209750
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC146587.10


