

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146586.12 - phase: 0 /pseudo
(840 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
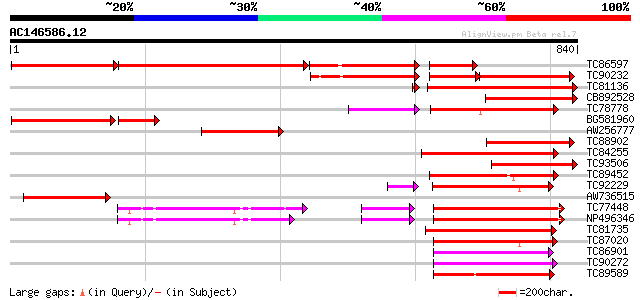
Score E
Sequences producing significant alignments: (bits) Value
TC86597 weakly similar to GP|9837280|gb|AAG00510.1| leaf senesce... 436 0.0
TC90232 weakly similar to PIR|T45697|T45697 hypothetical protein... 208 e-136
TC81136 weakly similar to GP|8778538|gb|AAF79546.1| F22G5.7 {Ara... 351 6e-98
CB892528 weakly similar to PIR|E96647|E966 hypothetical protein ... 260 1e-69
TC78778 weakly similar to PIR|S71277|S71277 serine/threonine-spe... 200 2e-65
BG581960 similar to PIR|A96557|A96 probable receptor protein kin... 151 2e-52
AW256777 weakly similar to PIR|T01269|T01 serine/threonine-speci... 202 3e-52
TC88902 weakly similar to GP|9294681|dbj|BAB03047.1 receptor-lik... 189 4e-48
TC84255 similar to GP|11072027|gb|AAG28906.1 F12A21.14 {Arabidop... 173 2e-43
TC93506 weakly similar to PIR|T45697|T45697 hypothetical protein... 171 1e-42
TC89452 similar to PIR|H84787|H84787 probable receptor-like prot... 168 7e-42
TC92229 similar to PIR|B86369|B86369 hypothetical protein AAC980... 150 1e-38
AW736515 weakly similar to GP|9837280|gb|A leaf senescence-assoc... 155 8e-38
TC77448 SYMRK; MtSYMRK [Medicago truncatula]; nod region linked ... 148 5e-37
NP496346 NP496346|AJ418371.1|CAD10810.1 nod region linked recept... 148 5e-37
TC81735 similar to GP|10998537|gb|AAG25966.1 cytokinin-regulated... 148 7e-36
TC87020 similar to GP|13447449|gb|AAK21965.1 receptor protein ki... 146 4e-35
TC86901 similar to GP|13937147|gb|AAK50067.1 AT5g13160/T19L5_120... 144 1e-34
TC90272 similar to PIR|T45686|T45686 receptor-like protein kinas... 144 1e-34
TC89589 similar to GP|14274168|emb|CAC40002. Arabidopsis thalian... 143 3e-34
>TC86597 weakly similar to GP|9837280|gb|AAG00510.1| leaf
senescence-associated receptor-like protein kinase
{Phaseolus vulgaris}, partial (46%)
Length = 2233
Score = 436 bits (1121), Expect(4) = 0.0
Identities = 218/282 (77%), Positives = 238/282 (84%)
Frame = +3
Query: 161 QYKDDVYDRMWFSYELIDWRRLSTSLNNDHLVQNIYKPPTIVMSTAATPVNASAPLQFHW 220
+YKDDV+DR+WF Y IDW RLSTSLNND LVQN Y+PP VMSTA TP+NASAPLQF W
Sbjct: 693 RYKDDVFDRIWFPYGFIDWARLSTSLNNDDLVQNDYEPPATVMSTAVTPLNASAPLQFEW 872
Query: 221 SSNNENDQYYLYIHFNEVEELAANETREFNITVNDKLWFGPVTPIYRTPDLIFSTEPLRR 280
++N NDQYY Y HFNEVE+LA NETR FN++VN GPV P YR IFS PL
Sbjct: 873 GTDNVNDQYYTYFHFNEVEKLAENETRAFNLSVNGNHRHGPVIPGYRETYTIFSPTPLTG 1052
Query: 281 AETYQISLSKTKNSTLPPILNAFEIYMAKDFSQLETQQDDVDNITNIKNAYGVTRNWQGD 340
A++YQISL+KT+NSTLPPILNAFEIY KDFSQ ET QDDVD ITNIKNAYG+TRNW+GD
Sbjct: 1053AKSYQISLTKTENSTLPPILNAFEIYKVKDFSQSETHQDDVDTITNIKNAYGLTRNWEGD 1232
Query: 341 PCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLN 400
PCAPVNY+WEGLNCS D NN PRI SLNLSSSGLTGEISSSISKLTMLQYLDLSNNSL+
Sbjct: 1233PCAPVNYVWEGLNCSI-DGNNIPRIISLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLS 1409
Query: 401 GPLPDFLIQLRSLQVLNVGKNNLTGLVPSELLERSKTGSLSL 442
GPLPDFL+QLRSL+VLNVGKN LTGLVPS LLERSKTGSL L
Sbjct: 1410GPLPDFLMQLRSLKVLNVGKNKLTGLVPSGLLERSKTGSLLL 1535
Score = 226 bits (576), Expect(4) = 0.0
Identities = 126/163 (77%), Positives = 133/163 (81%)
Frame = +2
Query: 444 **SRPL*KGIMQKEEEFVCTTYSIVFSNDCNRLDFSWILDIQKKETSYNYVFKL*E*GFN 503
**S L* IMQKEEE VCTTY I+FS CN LDFSWI D+QK N K * GFN
Sbjct: 1544 **SWSL*D*IMQKEEEIVCTTYCIIFSIYCNPLDFSWISDVQKT----NSYIKF*GKGFN 1711
Query: 504 KIKTSKIQLY*NCQYY**LQNYYWRRRIWKSLLWHSTRSN*SRC*DAFTIINARL*GI*S 563
+IKTSK+QL+*N QYY**LQNYYWRRRIWKSLLWHST SN SRC*DAFTIINARL*GI
Sbjct: 1712 EIKTSKVQLH*NSQYY**LQNYYWRRRIWKSLLWHSTGSNSSRC*DAFTIINARL*GISI 1891
Query: 564 RGTTSNSCSS*KFGLPCWIL**R*NQSINL*IHGQWKSPTTFI 606
RGTTSN+CSS*KFGLP WIL**R*NQSI+L*IHG WKS TT I
Sbjct: 1892 RGTTSNNCSS*KFGLPHWIL**R*NQSIDL*IHG*WKSATTII 2020
Score = 166 bits (420), Expect(4) = 0.0
Identities = 110/161 (68%), Positives = 121/161 (74%), Gaps = 2/161 (1%)
Frame = +1
Query: 3 QDSLA*IVVYLNI*AILIRTQT*ITFQMPNS*IAV*VRRYCLQTMF-DGI*NM*EAFQVE 61
QDSLA*IVVYL I* IL R Q *IT+QMPNS*I V* R +Q F *NM*EAFQVE
Sbjct: 148 QDSLA*IVVYLKI*TILHRRQA*ITYQMPNS*ILV*ARG*HVQISFLYSN*NM*EAFQVE 327
Query: 62 *EIVTE*M*HLVLNI*SELLSIMVTMMI*MTLHNLIFILELMFGIL*SSP-MHHV*DSMR 120
*E VT *+* +VLNI SELL M TM+I* + HNLIF LELM GI *S P MHH S +
Sbjct: 328 *ETVTT*V*QVVLNILSELLFFMETMII*TSHHNLIFTLELMCGIQ*SCPPMHHSSHSGK 507
Query: 121 SFTVHHKITYNHVLLTQAKGLHSFRL*N*GL*TMKLTSHLQ 161
SFT+ ITYNHVLLTQ KGLHSF+L*N*GL*T++L SH Q
Sbjct: 508 SFTLRR*ITYNHVLLTQEKGLHSFQL*N*GL*TIQLMSHTQ 630
Score = 132 bits (333), Expect(4) = 0.0
Identities = 60/70 (85%), Positives = 66/70 (93%)
Frame = +3
Query: 623 VENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLS 682
VENS +LNWNERLNIAVDAA+GL+YLHNGCKPP MHRDLKPSNILLD++MHAKI DFGLS
Sbjct: 2022 VENSKILNWNERLNIAVDAAYGLEYLHNGCKPPIMHRDLKPSNILLDDDMHAKIFDFGLS 2201
Query: 683 RAFDNDIDSH 692
RAF ND+DSH
Sbjct: 2202 RAFGNDVDSH 2231
>TC90232 weakly similar to PIR|T45697|T45697 hypothetical protein F18L15.120
- Arabidopsis thaliana, partial (26%)
Length = 1272
Score = 208 bits (530), Expect(3) = e-136
Identities = 101/140 (72%), Positives = 120/140 (85%)
Frame = +1
Query: 697 PAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALVRASGESIHILQWVTPIVE 756
PAGT GY DP++QRTGNTNKKNDIYSFGI+L ELITG+KAL +ASGE++HIL+WV PIVE
Sbjct: 817 PAGTLGYADPEYQRTGNTNKKNDIYSFGIILFELITGQKALTKASGENLHILEWVIPIVE 996
Query: 757 RGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAELKECLSLDMVHR 816
GDI++++D+RLQG+F INSAWKVVEIAMS TSP VERPDMS+IL ELKECLSLDMV R
Sbjct: 997 GGDIQNVVDSRLQGEFSINSAWKVVEIAMSCTSPDVVERPDMSEILVELKECLSLDMVQR 1176
Query: 817 NNGRERAIVELTSLNIASDT 836
N G RA+ EL S+ S++
Sbjct: 1177NCGGTRAMDELVSVATISES 1236
Score = 173 bits (438), Expect(3) = e-136
Identities = 101/161 (62%), Positives = 117/161 (71%)
Frame = +2
Query: 446 SRPL*KGIMQKEEEFVCTTYSIVFSNDCNRLDFSWILDIQKKETSYNYVFKL*E*GFNKI 505
SR L KG+MQK E CT Y I F CN LD S IL+IQK ++ + F L E F I
Sbjct: 122 SRSLYKGLMQK*EH-CCTNYWITFRISCNPLDLSCILEIQKTKS---WTF*LQEERFTGI 289
Query: 506 KTSKIQLY*NCQYY**LQNYYWRRRIWKSLLWHSTRSN*SRC*DAFTIINARL*GI*SRG 565
T IQL+*N QY+**LQNYYWRRRIWKSL W+ T+ N S C*DAFTI+NARL* I RG
Sbjct: 290 HT*SIQLH*NSQYH**LQNYYWRRRIWKSLPWNFTKQNSSCC*DAFTILNARL*RISIRG 469
Query: 566 TTSNSCSS*KFGLPCWIL**R*NQSINL*IHGQWKSPTTFI 606
TS++CSS KFGL W+L**R NQSI+L*I+G+WKSPT FI
Sbjct: 470 ATSSNCSSSKFGLTHWVL**RGNQSIDL*IYGKWKSPTAFI 592
Score = 144 bits (362), Expect(3) = e-136
Identities = 65/75 (86%), Positives = 71/75 (94%)
Frame = +3
Query: 623 VENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLS 682
VENSN+LNWNERLNIAVDAA GLDY+HNGCKPP +HRDLKPSNILLD+NMHAKI+DFGLS
Sbjct: 594 VENSNILNWNERLNIAVDAAQGLDYMHNGCKPPILHRDLKPSNILLDDNMHAKISDFGLS 773
Query: 683 RAFDNDIDSHISTRP 697
RAF ND+DSHIST P
Sbjct: 774 RAFGNDVDSHISTGP 818
>TC81136 weakly similar to GP|8778538|gb|AAF79546.1| F22G5.7 {Arabidopsis
thaliana}, partial (10%)
Length = 868
Score = 351 bits (901), Expect(2) = 6e-98
Identities = 169/221 (76%), Positives = 195/221 (87%)
Frame = +3
Query: 620 LHAVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADF 679
++ +ENSN+LNW ERLNIAVD A+GLDYLHNGCKPP MHRDLKPSNILLDEN+HAKIADF
Sbjct: 24 IYQMENSNVLNWTERLNIAVDTAYGLDYLHNGCKPPIMHRDLKPSNILLDENLHAKIADF 203
Query: 680 GLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALVR 739
GLSRAF ND SHISTRPAGTFGY DP+FQR+GNTNKKNDI+SFGI+L ELITGKKAL R
Sbjct: 204 GLSRAFGNDDASHISTRPAGTFGYADPEFQRSGNTNKKNDIFSFGIILFELITGKKALER 383
Query: 740 ASGESIHILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMS 799
+ E+IHILQWV PI++ G+I++I+D+RLQG+F INSAWKVVEIAMS S VERPD++
Sbjct: 384 SYEENIHILQWVVPIIKAGNIQNIMDSRLQGEFSINSAWKVVEIAMSCVSQNAVERPDIN 563
Query: 800 QILAELKECLSLDMVHRNNGRERAIVELTSLNIASDTIPLA 840
QIL EL ECLSL+MV RNNGRERAIVE+TS+N+ S T PLA
Sbjct: 564 QILVELNECLSLEMVQRNNGRERAIVEITSVNLGSQTAPLA 686
Score = 25.8 bits (55), Expect(2) = 6e-98
Identities = 10/11 (90%), Positives = 10/11 (90%)
Frame = +1
Query: 597 GQWKSPTTFIR 607
GQWKS TTFIR
Sbjct: 1 GQWKSTTTFIR 33
>CB892528 weakly similar to PIR|E96647|E966 hypothetical protein F19K23.5
[imported] - Arabidopsis thaliana, partial (7%)
Length = 549
Score = 260 bits (665), Expect = 1e-69
Identities = 133/135 (98%), Positives = 134/135 (98%)
Frame = -1
Query: 706 PKFQRTGNTNKKNDIYSFGIVLLELITGKKALVRASGESIHILQWVTPIVERGDIRSIID 765
PKFQR GNTN+KNDIYSFGIVLLELITGKKALVRASGESIHILQWVTPIVERGDIRSIID
Sbjct: 549 PKFQRPGNTNRKNDIYSFGIVLLELITGKKALVRASGESIHILQWVTPIVERGDIRSIID 370
Query: 766 ARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAELKECLSLDMVHRNNGRERAIV 825
ARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAELKECLSLDMVHRNNGRERAIV
Sbjct: 369 ARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAELKECLSLDMVHRNNGRERAIV 190
Query: 826 ELTSLNIASDTIPLA 840
ELTSLNIASDTIPLA
Sbjct: 189 ELTSLNIASDTIPLA 145
>TC78778 weakly similar to PIR|S71277|S71277 serine/threonine-specific
receptor protein kinase (EC 2.7.1.-) - Arabidopsis
thaliana, partial (23%)
Length = 1808
Score = 200 bits (509), Expect(2) = 2e-65
Identities = 99/201 (49%), Positives = 144/201 (71%), Gaps = 12/201 (5%)
Frame = +3
Query: 624 ENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSR 683
++S+ L+W RL IA+DAA GLDYLH+GCKPP +HRD+K +NILL+E++ AKIADFGLS+
Sbjct: 990 KSSHCLSWERRLQIAIDAAEGLDYLHHGCKPPIIHRDVKSANILLNEDLEAKIADFGLSK 1169
Query: 684 AFDN-DIDSHIST-----------RPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELI 731
F N DI + ST GT GY+DP++ + N+K+DIYS+GIVLLELI
Sbjct: 1170 VFKNSDIQNADSTLIHVDVSGEKSAIMGTMGYLDPEYYKLQTLNEKSDIYSYGIVLLELI 1349
Query: 732 TGKKALVRASGESIHILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPI 791
TG A+++ S HIL++V P + + D+ +ID RL+GKFD++S WKV+ +A++ T+
Sbjct: 1350 TGLPAVIKGK-PSKHILEFVRPRLNKDDLSKVIDPRLEGKFDVSSGWKVLGLAIACTAST 1526
Query: 792 EVERPDMSQILAELKECLSLD 812
++RP MS +LAELK+C+ ++
Sbjct: 1527 SIQRPTMSVVLAELKQCVRME 1589
Score = 68.6 bits (166), Expect(2) = 2e-65
Identities = 43/105 (40%), Positives = 59/105 (55%)
Frame = +2
Query: 503 NKIKTSKIQLY*NCQYY**LQNYYWRRRIWKSLLWHSTRSN*SRC*DAFTIINARL*GI* 562
+ IK I L * ++ L N +W+ RIWK L W R *S DAF I ++R GI
Sbjct: 674 SSIKEVAIHLC*GVGHHQQLGNSHWKGRIWKCLQWSDERWQ*SCSQDAFCIFSSRAKGIS 853
Query: 563 SRGTTSNSCSS*KFGLPCWIL**R*NQSINL*IHGQWKSPTTFIR 607
+ G T + SS KFG+ W+L**R* ++*++GQWK +R
Sbjct: 854 N*G*TFDDSSSQKFGILHWLL**R*QDGTDI*VYGQWKPKRKSLR 988
>BG581960 similar to PIR|A96557|A96 probable receptor protein kinase
[imported] - Arabidopsis thaliana, partial (12%)
Length = 825
Score = 151 bits (381), Expect(2) = 2e-52
Identities = 107/157 (68%), Positives = 117/157 (74%), Gaps = 3/157 (1%)
Frame = +1
Query: 3 QDSLA*IVVYLNI*AILIRTQT*ITFQMPNS*-IAV*VRRYCLQTMF-DGI*NM*EAFQV 60
QDSLA*IV L + IL Q *ITFQMPNS I V*VR Y L+ F D * * AFQV
Sbjct: 79 QDSLA*IVGDLKMRTILRWRQA*ITFQMPNSSSIKV*VRAYNLRKSFLDRN*RK*GAFQV 258
Query: 61 E*EIVTE*M*HLVLNI*SELLSIMVTMMI*MTLHNLIFILELMFGIL*S-SPMHHV*DSM 119
E*E VT+*M* +VLNI* ELLSIM TM+I*M HNLIFILELM GI *S SPMHH S
Sbjct: 259 E*ETVTK*M*QVVLNI**ELLSIMETMII*MNHHNLIFILELMCGIQ*SCSPMHHTSLSR 438
Query: 120 RSFTVHHKITYNHVLLTQAKGLHSFRL*N*GL*TMKL 156
R FT+H IT NHVLLTQAKGLHSF+ *N*GL*T+++
Sbjct: 439 RLFTLHQ*IT*NHVLLTQAKGLHSFQP*N*GL*TIRI 549
Score = 74.3 bits (181), Expect(2) = 2e-52
Identities = 38/64 (59%), Positives = 48/64 (74%), Gaps = 2/64 (3%)
Frame = +3
Query: 161 QYKDDVYDRMWFSYELID-WRRLSTSLNNDHL-VQNIYKPPTIVMSTAATPVNASAPLQF 218
+Y+ DVYDR W +L RRL+TS+ N +L + NIY+P +IVMSTA TPVNASAPLQF
Sbjct: 633 RYRKDVYDRNWMPCKLPSVCRRLNTSVKNVNLPIPNIYEPTSIVMSTAVTPVNASAPLQF 812
Query: 219 HWSS 222
HW +
Sbjct: 813 HWDA 824
>AW256777 weakly similar to PIR|T01269|T01 serine/threonine-specific protein
kinase (EC 2.7.1.-) F27F23.1 - Arabidopsis thaliana,
partial (13%)
Length = 373
Score = 202 bits (515), Expect = 3e-52
Identities = 101/122 (82%), Positives = 105/122 (85%)
Frame = +3
Query: 284 YQISLSKTKNSTLPPILNAFEIYMAKDFSQLETQQDDVDNITNIKNAYGVTRNWQGDPCA 343
Y +LSKT NSTLPPILNA E+Y K+FSQ ETQQDDVD + NIK AYGV RNWQGDPC
Sbjct: 9 YIFTLSKTDNSTLPPILNAVEVYKVKNFSQSETQQDDVDTMRNIKKAYGVARNWQGDPCG 188
Query: 344 PVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPL 403
PVNYMWEGLNCS D NN PRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNG L
Sbjct: 189 PVNYMWEGLNCSL-DGNNIPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGSL 365
Query: 404 PD 405
PD
Sbjct: 366 PD 371
>TC88902 weakly similar to GP|9294681|dbj|BAB03047.1 receptor-like protein
kinase {Arabidopsis thaliana}, partial (7%)
Length = 748
Score = 189 bits (480), Expect = 4e-48
Identities = 92/130 (70%), Positives = 110/130 (83%)
Frame = +1
Query: 707 KFQRTGNTNKKNDIYSFGIVLLELITGKKALVRASGESIHILQWVTPIVERGDIRSIIDA 766
+FQRTGNTNKKNDIYSFGI+L ELITG+KAL++AS ++IHIL+WV PIVE GDI++++D
Sbjct: 73 RFQRTGNTNKKNDIYSFGIILFELITGQKALIKASEKTIHILEWVIPIVEGGDIQNVVDL 252
Query: 767 RLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAELKECLSLDMVHRNNGRERAIVE 826
RLQG+F INSAWK VEIAMS TSP +ERPDMSQIL +LKECLSLDMV RN G RAI +
Sbjct: 253 RLQGEFSINSAWKAVEIAMSCTSPNAIERPDMSQILVDLKECLSLDMVQRNYGSTRAIDD 432
Query: 827 LTSLNIASDT 836
+ SL S+T
Sbjct: 433 MISLATVSET 462
>TC84255 similar to GP|11072027|gb|AAG28906.1 F12A21.14 {Arabidopsis
thaliana}, partial (20%)
Length = 743
Score = 173 bits (439), Expect = 2e-43
Identities = 86/204 (42%), Positives = 124/204 (60%), Gaps = 1/204 (0%)
Frame = +2
Query: 610 HFQNFETNAYLHAVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLD 669
+ N ++H + L+W RL IA DAA GL+YLH GC P +HRD+K SNILLD
Sbjct: 32 YMHNGTLRDHIHECSSEKRLDWLTRLRIAEDAAKGLEYLHTGCNPSIIHRDVKTSNILLD 211
Query: 670 ENMHAKIADFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLE 729
NM AK++DFGLSR + D+ +HIS+ GT GY+DP++ +K+D+YSFG+VLLE
Sbjct: 212 INMRAKVSDFGLSRLAEEDL-THISSVAKGTVGYLDPEYYANQQLTEKSDVYSFGVVLLE 388
Query: 730 LITGKKALVRAS-GESIHILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSST 788
LI GKK + G ++I+ W ++ +GDI SI+D L G S W+V EIAM
Sbjct: 389 LICGKKPVSPEDYGPEMNIVHWARSLIRKGDIISIMDPLLIGNVKTESIWRVAEIAMQCV 568
Query: 789 SPIEVERPDMSQILAELKECLSLD 812
P RP M +++ +++ ++
Sbjct: 569 EPHGASRPRMQEVILAIQDASKIE 640
>TC93506 weakly similar to PIR|T45697|T45697 hypothetical protein F18L15.120
- Arabidopsis thaliana, partial (8%)
Length = 488
Score = 171 bits (433), Expect = 1e-42
Identities = 91/127 (71%), Positives = 104/127 (81%), Gaps = 1/127 (0%)
Frame = +3
Query: 715 NKKNDIYSFGIVLLELITGKKALVRASGESIHILQWVTPIVERGDIRSIIDARLQGKFDI 774
NKKNDIYSFGI+L LITGKKALVR SGESIHILQWV PIV+RGDI++I+D +LQG+F+I
Sbjct: 3 NKKNDIYSFGIILFVLITGKKALVRESGESIHILQWVIPIVKRGDIQNIVDKKLQGEFNI 182
Query: 775 NSAWKVVEIAMSSTSPIEVERPDMSQILAELKECLSLDMVHRNNGRERAIVELTSLNI-A 833
+SAWKVVEIAMS S ERPD+SQILAELKECLSLDMV NNG RA EL S+ +
Sbjct: 183 SSAWKVVEIAMSCISQTVSERPDISQILAELKECLSLDMVQSNNGSMRARDELVSIALHV 362
Query: 834 SDTIPLA 840
S+T LA
Sbjct: 363 SETTILA 383
>TC89452 similar to PIR|H84787|H84787 probable receptor-like protein kinase
[imported] - Arabidopsis thaliana, partial (21%)
Length = 873
Score = 168 bits (426), Expect = 7e-42
Identities = 85/195 (43%), Positives = 128/195 (65%), Gaps = 5/195 (2%)
Frame = +1
Query: 623 VENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLS 682
+E+ +NW +RL IA DAA G++YLH GC P +HRDLK SNILLD M AK++DFGLS
Sbjct: 13 LEHGRSINWIKRLVIAEDAAKGIEYLHTGCVPVVIHRDLKTSNILLDRQMRAKVSDFGLS 192
Query: 683 RAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALVRASG 742
+ + + SH+S+ GT GY+DP++ + K+D+YSFG++LLELI+G++A+ S
Sbjct: 193 KLAVDGV-SHVSSIVRGTVGYLDPEYYISQQLTDKSDVYSFGVILLELISGQEAI---SN 360
Query: 743 ESI-----HILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPD 797
ES +I+QW +E GDI+ IID L +D+ S WK+ E A+ P RP
Sbjct: 361 ESFGLHCRNIVQWAKLHIESGDIQGIIDPLLGSNYDLQSMWKIAEKALMCVQPHGDMRPS 540
Query: 798 MSQILAELKECLSLD 812
+S++L E+++ +S++
Sbjct: 541 ISEVLKEIQDAISIE 585
>TC92229 similar to PIR|B86369|B86369 hypothetical protein AAC98010.1
[imported] - Arabidopsis thaliana, partial (44%)
Length = 1347
Score = 150 bits (378), Expect(2) = 1e-38
Identities = 76/185 (41%), Positives = 119/185 (64%), Gaps = 6/185 (3%)
Frame = +2
Query: 627 NMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFD 686
N+L+W +R+ IA+ AA GL YLH GC P +HRD+K SNILLD++ A++ADFGL+R D
Sbjct: 467 NVLDWPKRMKIAIGAARGLAYLHEGCNPKIIHRDIKSSNILLDDSYEAQVADFGLARLTD 646
Query: 687 NDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKAL--VRASGES 744
D ++H+STR GTFGY+ P++ +G ++D++SFG+VLLEL+TG+K + + G+
Sbjct: 647 -DTNTHVSTRVMGTFGYMAPEYATSGKLTDRSDVFSFGVVLLELVTGRKPVDPTQPVGDE 823
Query: 745 IHILQWVTPI----VERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQ 800
+++W PI +E GD + D RL ++ + ++++E A + +RP M Q
Sbjct: 824 -SLVEWARPILLRAIETGDFSELADPRLHRQYIDSEMFRMIEAAAACIRHSAPKRPRMVQ 1000
Query: 801 ILAEL 805
I L
Sbjct: 1001IARAL 1015
Score = 28.9 bits (63), Expect(2) = 1e-38
Identities = 20/46 (43%), Positives = 27/46 (58%)
Frame = +1
Query: 560 GI*SRGTTSNSCSS*KFGLPCWIL**R*NQSINL*IHGQWKSPTTF 605
GI SRG SCSS FG+ W+L +S +L*+ W+S +TF
Sbjct: 313 GIQSRG*YY*SCSSSTFGIVDWLLYS*ATKSPHL*VCS*WES*STF 450
>AW736515 weakly similar to GP|9837280|gb|A leaf senescence-associated
receptor-like protein kinase {Phaseolus vulgaris},
partial (9%)
Length = 397
Score = 155 bits (391), Expect = 8e-38
Identities = 94/130 (72%), Positives = 104/130 (79%), Gaps = 1/130 (0%)
Frame = +1
Query: 21 RTQT*ITFQMPNS*IAV*VRRYCLQTMF-DGI*NM*EAFQVE*EIVTE*M*HLVLNI*SE 79
RTQ *ITFQMPNS*I V*VR Y LQ + + *NM* F VE*EIVT+*M* ++LNI*SE
Sbjct: 4 RTQA*ITFQMPNS*IQV*VRGYLLQRLLLNNN*NM*GVFLVE*EIVTK*M*QVILNI*SE 183
Query: 80 LLSIMVTMMI*MTLHNLIFILELMFGIL*SSPMHHV*DSMRSFTVHHKITYNHVLLTQAK 139
LSIM TMMI* + HNLIFIL LMFGI *SS ++H +RSFT+HHKITYNHVLLTQA
Sbjct: 184 RLSIMETMMI*TSHHNLIFILALMFGIQ*SSQIYHSSQPVRSFTLHHKITYNHVLLTQAT 363
Query: 140 GLHSFRL*N* 149
GLHSF L*N*
Sbjct: 364 GLHSFYL*N* 393
>TC77448 SYMRK; MtSYMRK [Medicago truncatula]; nod region linked receptor
kinase [Medicago truncatula]
Length = 3568
Score = 148 bits (374), Expect(2) = 5e-37
Identities = 73/195 (37%), Positives = 121/195 (61%), Gaps = 1/195 (0%)
Frame = +1
Query: 628 MLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDN 687
+L+W RL+IA+ AA GL YLH +HRD+K SNILLD++M AK+ADFG S+
Sbjct: 2440 ILDWPTRLSIALGAARGLAYLHTFPGRSVIHRDVKSSNILLDQSMCAKVADFGFSKYAPQ 2619
Query: 688 DIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKAL-VRASGESIH 746
+ DS++S GT GY+DP++ +T ++K+D++SFG+VLLE+++G++ L ++
Sbjct: 2620 EGDSYVSLEVRGTAGYLDPEYYKTQQLSEKSDVFSFGVVLLEIVSGREPLNIKRPRIEWS 2799
Query: 747 ILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAELK 806
+++W P + + I+D ++G + + W+VVE+A+ P RP M I+ EL+
Sbjct: 2800 LVEWAKPYIRASKVDEIVDPGIKGGYHAEALWRVVEVALQCLEPYSTYRPCMVDIVRELE 2979
Query: 807 ECLSLDMVHRNNGRE 821
+ L ++ NN E
Sbjct: 2980 DALIIE----NNASE 3012
Score = 96.7 bits (239), Expect = 3e-20
Identities = 82/293 (27%), Positives = 141/293 (47%), Gaps = 12/293 (4%)
Frame = +1
Query: 160 LQYKDDVYDRMWFSYEL---IDWRRLSTSLNNDHLVQNIYKPPTIVMSTAATPVNASAPL 216
+++ DD DR+W E LS +++N L ++ PP V+ TA T L
Sbjct: 976 IRFPDDQNDRIWKRKETSTPTSALPLSFNVSNVDLKDSV-TPPLQVLQTALTHPER---L 1143
Query: 217 QF-HWSSNNENDQYYLYIHFNEVEELAANETREFNITVNDKLWFGPVTPIYRTPDLIFST 275
+F H ++ +Y +++HF E+ R F+I +N+++ + ++
Sbjct: 1144 EFVHDGLETDDYEYSVFLHFLELNGTVRAGQRVFDIYLNNEIKKEKFDVLAGGSKNSYTA 1323
Query: 276 EPLRRAETYQISLSKTKNSTLPPILNAFEIYMAKDFSQLETQQDDVDNITNIKNAY---- 331
+ + I+L K S P+LNA+EI A+ + + ET Q D++ I ++
Sbjct: 1324 LNISANGSLNITLVKASGSEFGPLLNAYEILQARSWIE-ETNQKDLEVIQKMREELLLHN 1500
Query: 332 ---GVTRNWQGDPCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTM 388
+W GDPC + + W+G+ C DD IT L+LSS+ L G I S ++K+T
Sbjct: 1501 QENEALESWSGDPC--MIFPWKGITC--DDSTGSSIITKLDLSSNNLKGAIPSIVTKMTN 1668
Query: 389 LQYLDLSNNSLNGPLPDFLIQLRSLQV-LNVGKNNLTGLVPSELLERSKTGSL 440
LQ L+LS+N + P F SL + L++ N+L+G +P ++ SL
Sbjct: 1669 LQILNLSHNQFDMLFPSF--PPSSLLISLDLSYNDLSGWLPESIISLPHLKSL 1821
Score = 25.0 bits (53), Expect(2) = 5e-37
Identities = 26/78 (33%), Positives = 38/78 (48%)
Frame = +3
Query: 522 LQNYYWRRRIWKSLLWHSTRSN*SRC*DAFTIINARL*GI*SRGTTSNSCSS*KFGLPCW 581
+QN+ RRIW L HS R + S A IN+ I* + S ++*+ G
Sbjct: 2163 VQNFNR*RRIWLCLQGHSRRWSRSGSESAVIHINSGNPRI***AKPTFSYTT*EPGASSG 2342
Query: 582 IL**R*NQSINL*IHGQW 599
+L** *+ + + H QW
Sbjct: 2343 LL**V*STNSRVSFHVQW 2396
>NP496346 NP496346|AJ418371.1|CAD10810.1 nod region linked receptor kinase
[Medicago truncatula]
Length = 2706
Score = 148 bits (374), Expect(2) = 5e-37
Identities = 73/195 (37%), Positives = 121/195 (61%), Gaps = 1/195 (0%)
Frame = +1
Query: 628 MLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDN 687
+L+W RL+IA+ AA GL YLH +HRD+K SNILLD++M AK+ADFG S+
Sbjct: 1996 ILDWPTRLSIALGAARGLAYLHTFPGRSVIHRDVKSSNILLDQSMCAKVADFGFSKYAPQ 2175
Query: 688 DIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKAL-VRASGESIH 746
+ DS++S GT GY+DP++ +T ++K+D++SFG+VLLE+++G++ L ++
Sbjct: 2176 EGDSYVSLEVRGTAGYLDPEYYKTQQLSEKSDVFSFGVVLLEIVSGREPLNIKRPRIEWS 2355
Query: 747 ILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAELK 806
+++W P + + I+D ++G + + W+VVE+A+ P RP M I+ EL+
Sbjct: 2356 LVEWAKPYIRASKVDEIVDPGIKGGYHAEALWRVVEVALQCLEPYSTYRPCMVDIVRELE 2535
Query: 807 ECLSLDMVHRNNGRE 821
+ L ++ NN E
Sbjct: 2536 DALIIE----NNASE 2568
Score = 104 bits (259), Expect = 2e-22
Identities = 82/273 (30%), Positives = 134/273 (49%), Gaps = 11/273 (4%)
Frame = +1
Query: 160 LQYKDDVYDRMWFSYEL---IDWRRLSTSLNNDHLVQNIYKPPTIVMSTAATPVNASAPL 216
+++ DD DR+W E LS +++N L ++ PP V+ TA T L
Sbjct: 601 IRFPDDQNDRIWKRKETSTPTSALPLSFNVSNVDLKDSV-TPPLQVLQTALTHPER---L 768
Query: 217 QF-HWSSNNENDQYYLYIHFNEVEELAANETREFNITVNDKLWFGPVTPIYRTPDLIFST 275
+F H ++ +Y +++HF E+ R F+I +N+++ + ++
Sbjct: 769 EFVHDGLETDDYEYSVFLHFLELNGTVRAGQRVFDIYLNNEIKKEKFDVLAGGSKNSYTA 948
Query: 276 EPLRRAETYQISLSKTKNSTLPPILNAFEIYMAKDFSQLETQQDDVDNITNIKNAY---- 331
+ + I+L K S P+LNA+EI A+ + + ET Q D++ I ++
Sbjct: 949 LNISANGSLNITLVKASGSEFGPLLNAYEILQARSWIE-ETNQKDLEVIQKMREELLLHN 1125
Query: 332 ---GVTRNWQGDPCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTM 388
+W GDPC + + W+G+ C DD IT L+LSS+ L G I S ++K+T
Sbjct: 1126QENEALESWSGDPC--MIFPWKGITC--DDSTGSSIITKLDLSSNNLKGAIPSIVTKMTN 1293
Query: 389 LQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKN 421
LQ LDLS N L+G LP+ +I L L+ L G N
Sbjct: 1294LQILDLSYNDLSGWLPESIISLPHLKSLYFGCN 1392
Score = 25.0 bits (53), Expect(2) = 5e-37
Identities = 26/78 (33%), Positives = 38/78 (48%)
Frame = +3
Query: 522 LQNYYWRRRIWKSLLWHSTRSN*SRC*DAFTIINARL*GI*SRGTTSNSCSS*KFGLPCW 581
+QN+ RRIW L HS R + S A IN+ I* + S ++*+ G
Sbjct: 1719 VQNFNR*RRIWLCLQGHSRRWSRSGSESAVIHINSGNPRI***AKPTFSYTT*EPGASSG 1898
Query: 582 IL**R*NQSINL*IHGQW 599
+L** *+ + + H QW
Sbjct: 1899 LL**V*STNSRVSFHVQW 1952
>TC81735 similar to GP|10998537|gb|AAG25966.1 cytokinin-regulated kinase 1
{Nicotiana tabacum}, partial (28%)
Length = 772
Score = 148 bits (374), Expect = 7e-36
Identities = 76/196 (38%), Positives = 123/196 (61%), Gaps = 3/196 (1%)
Frame = +2
Query: 617 NAYLHAVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKI 676
N +LH + S +++W+ R+ +A+DAA G++YLH +PP +HRD+K SNILLD AK+
Sbjct: 68 NDHLHKFQTSTIMSWSGRIKVALDAARGIEYLHKYAQPPIIHRDIKTSNILLDSKWVAKV 247
Query: 677 ADFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKA 736
+DFGLS D +SH+S AGT GY+DP++ R K+D+YSFG+VLLEL++G KA
Sbjct: 248 SDFGLSLMGPEDEESHLSLLAAGTVGYMDPEYYRLQYLTSKSDVYSFGVVLLELLSGYKA 427
Query: 737 LVR-ASGESIHILQWVTPIVERGDIRSIIDARL--QGKFDINSAWKVVEIAMSSTSPIEV 793
+ + +G +++ +V P + + +I I+D +L F+I + V +A
Sbjct: 428 IHKNENGVPRNVVDFVVPYIVQDEIHRILDTKLPPPTPFEIEAVTFVGYLACDCVRLEGR 607
Query: 794 ERPDMSQILAELKECL 809
+RP+MS ++ L++ L
Sbjct: 608 DRPNMSHVVNSLEKAL 655
>TC87020 similar to GP|13447449|gb|AAK21965.1 receptor protein kinase PERK1
{Brassica napus}, partial (39%)
Length = 1150
Score = 146 bits (368), Expect = 4e-35
Identities = 74/188 (39%), Positives = 121/188 (64%), Gaps = 5/188 (2%)
Frame = +1
Query: 629 LNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDND 688
++W+ RL IA+ +A GL YLH C P +HRD+K +NILLD AK+ADFGL++ +D
Sbjct: 28 MDWSTRLRIALGSAKGLAYLHEDCHPKIIHRDIKAANILLDFKFEAKVADFGLAK-IASD 204
Query: 689 IDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALVR-ASGESIHI 747
+++H+STR GTFGY+ P++ +G K+D++S+G++LLEL+TG++ + + + +
Sbjct: 205 LNTHVSTRVMGTFGYLAPEYAASGKLTDKSDVFSYGVMLLELLTGRRPVDKDQTYMDDSL 384
Query: 748 LQWVTPI----VERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILA 803
++W P+ +E ++ S+ID RLQ FD N ++V A + T RP MSQ++
Sbjct: 385 VEWARPLLMRALEEDNLDSLIDPRLQNDFDPNEMTRMVACAAACTRHSAKRRPKMSQVVR 564
Query: 804 ELKECLSL 811
L+ +SL
Sbjct: 565 ALEGDVSL 588
>TC86901 similar to GP|13937147|gb|AAK50067.1 AT5g13160/T19L5_120
{Arabidopsis thaliana}, partial (83%)
Length = 2028
Score = 144 bits (363), Expect = 1e-34
Identities = 73/180 (40%), Positives = 108/180 (59%), Gaps = 3/180 (1%)
Frame = +1
Query: 629 LNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDND 688
L+WN R+ IA AA GL+YLH+ PP ++RD K SNILLDE H K++DFGL++
Sbjct: 817 LDWNTRMKIAAGAAKGLEYLHDKANPPVIYRDFKSSNILLDEGYHPKLSDFGLAKLGPVG 996
Query: 689 IDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKAL--VRASGESIH 746
SH+STR GT+GY P++ TG K+D+YSFG+V LELITG+KA+ R GE +
Sbjct: 997 DKSHVSTRVMGTYGYCAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDSTRPHGEQ-N 1173
Query: 747 ILQWVTPIV-ERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAEL 805
++ W P+ +R + D RLQG++ + ++ + +A RP + ++ L
Sbjct: 1174LVTWARPLFNDRRKFSKLADPRLQGRYPMRGLYQALAVASMCIQEQAAARPLIGDVVTAL 1353
>TC90272 similar to PIR|T45686|T45686 receptor-like protein kinase homolog -
Arabidopsis thaliana, partial (58%)
Length = 941
Score = 144 bits (363), Expect = 1e-34
Identities = 73/184 (39%), Positives = 109/184 (58%), Gaps = 1/184 (0%)
Frame = +3
Query: 629 LNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDND 688
L+W ERL I + AA GL YLH G +HRD+K +NILLDEN+ AK+ADFGLS+
Sbjct: 210 LSWKERLEICIGAARGLHYLHTGYAKAVIHRDVKSANILLDENLMAKVADFGLSKTGPEI 389
Query: 689 IDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKAL-VRASGESIHI 747
+H+ST G+FGY+DP++ R +K+D+YSFG+VLLE++ + + E +++
Sbjct: 390 DQTHVSTAVKGSFGYLDPEYFRRQQLTEKSDVYSFGVVLLEVLCARPVIDPSLPRERVNL 569
Query: 748 LQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAELKE 807
+W ++G++ I+D L GK +S K E A + V+RP M +L L+
Sbjct: 570 AEWAMKWQKKGELARIVDPTLAGKIRPDSLRKFAETAEKCLADFGVDRPSMGDVLWNLEY 749
Query: 808 CLSL 811
L L
Sbjct: 750 ALQL 761
>TC89589 similar to GP|14274168|emb|CAC40002. Arabidopsis thaliana RKS1
cDNA, partial (36%)
Length = 814
Score = 143 bits (360), Expect = 3e-34
Identities = 73/180 (40%), Positives = 113/180 (62%), Gaps = 2/180 (1%)
Frame = +2
Query: 629 LNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDND 688
L+W R IA+ A GL YLH C P +HRD+K +NILLDE+ A + DFGL++ D+
Sbjct: 23 LDWTRRKRIALGTARGLVYLHEQCDPKIIHRDVKAANILLDEDFEAVVGDFGLAKLLDHR 202
Query: 689 IDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKAL--VRASGESIH 746
D+H++T GT G++ P++ TG +++K D++ +GI+LLELITG KAL RA+ +
Sbjct: 203 -DTHVTTAVRGTIGHIAPEYLSTGQSSEKTDVFGYGILLLELITGHKALDFGRAANQKGV 379
Query: 747 ILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAELK 806
+L WV + G + ++D L+G FDI ++V++A+ T RP MS++L L+
Sbjct: 380 MLDWVKKLHLEGKLSQMVDKDLKGNFDIVELGEMVQVALLCTQFNPSHRPKMSEVLKMLE 559
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.343 0.150 0.492
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,996,759
Number of Sequences: 36976
Number of extensions: 372531
Number of successful extensions: 5162
Number of sequences better than 10.0: 755
Number of HSP's better than 10.0 without gapping: 2360
Number of HSP's successfully gapped in prelim test: 216
Number of HSP's that attempted gapping in prelim test: 2085
Number of HSP's gapped (non-prelim): 3019
length of query: 840
length of database: 9,014,727
effective HSP length: 104
effective length of query: 736
effective length of database: 5,169,223
effective search space: 3804548128
effective search space used: 3804548128
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146586.12


