

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146586.11 - phase: 0 /pseudo
(291 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
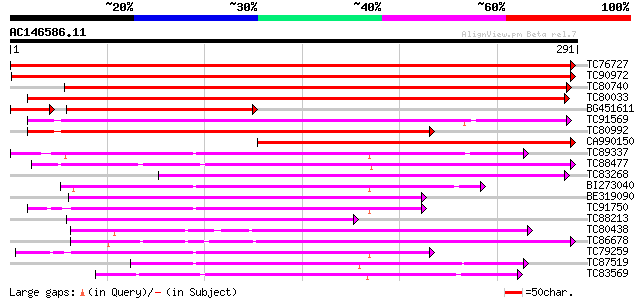
Score E
Sequences producing significant alignments: (bits) Value
TC76727 similar to GP|10638955|emb|CAB81548. putative proline-ri... 584 e-167
TC90972 similar to GP|10638955|emb|CAB81548. putative proline-ri... 454 e-128
TC80740 similar to GP|10177228|dbj|BAB10602. GDSL-motif lipase/h... 320 3e-88
TC80033 weakly similar to GP|21593518|gb|AAM65485.1 putative GDS... 238 3e-63
BG451611 homologue to GP|10638955|em putative proline-rich prote... 203 3e-61
TC91569 similar to GP|18252233|gb|AAL61949.1 putative APG protei... 224 2e-59
TC80992 weakly similar to GP|21593518|gb|AAM65485.1 putative GDS... 186 7e-48
CA990150 weakly similar to GP|10177228|db GDSL-motif lipase/hydr... 176 7e-45
TC89337 similar to GP|21592967|gb|AAM64916.1 putative GDSL-motif... 157 6e-39
TC88477 similar to GP|21592417|gb|AAM64368.1 lipase/hydrolase p... 153 7e-38
TC83268 weakly similar to GP|21592974|gb|AAM64923.1 proline-rich... 152 1e-37
BI273040 similar to GP|6721157|gb| putative GDSL-motif lipase/ac... 149 9e-37
BE319090 weakly similar to GP|15054386|gb family II lipase EXL3 ... 149 9e-37
TC91750 similar to GP|6721157|gb|AAF26785.1| putative GDSL-motif... 143 7e-35
TC88213 weakly similar to GP|15054386|gb|AAK30018.1 family II li... 139 1e-33
TC80438 similar to PIR|E96579|E96579 hypothetical protein T18A20... 139 1e-33
TC86678 similar to PIR|E96579|E96579 hypothetical protein T18A20... 139 1e-33
TC79259 similar to GP|6721157|gb|AAF26785.1| putative GDSL-motif... 130 8e-31
TC87519 weakly similar to GP|21592967|gb|AAM64916.1 putative GDS... 127 7e-30
TC83569 weakly similar to GP|6733842|emb|CAB69346.1 unnamed prot... 118 2e-27
>TC76727 similar to GP|10638955|emb|CAB81548. putative proline-rich protein
APG isolog {Cicer arietinum}, partial (92%)
Length = 1919
Score = 584 bits (1506), Expect = e-167
Identities = 290/290 (100%), Positives = 290/290 (100%)
Frame = -2
Query: 1 MMNMNSKETLVLLIVSCFLTCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPY 60
MMNMNSKETLVLLIVSCFLTCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPY
Sbjct: 1180 MMNMNSKETLVLLIVSCFLTCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPY 1001
Query: 61 GRDFTNKQPTGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASGYD 120
GRDFTNKQPTGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASGYD
Sbjct: 1000 GRDFTNKQPTGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASGYD 821
Query: 121 EKAATLNHAIPLSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTN 180
EKAATLNHAIPLSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTN
Sbjct: 820 EKAATLNHAIPLSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTN 641
Query: 181 PWINQAITVDQYSSYLLDSFTNFIKGVYGLGARKIGVTSLPPLGCLPAARTLFGYHENGC 240
PWINQAITVDQYSSYLLDSFTNFIKGVYGLGARKIGVTSLPPLGCLPAARTLFGYHENGC
Sbjct: 640 PWINQAITVDQYSSYLLDSFTNFIKGVYGLGARKIGVTSLPPLGCLPAARTLFGYHENGC 461
Query: 241 VARINTDAQGFNKKVSSAASNLQKQLPGLKIVIFDIYKPLYDLVQNPSNF 290
VARINTDAQGFNKKVSSAASNLQKQLPGLKIVIFDIYKPLYDLVQNPSNF
Sbjct: 460 VARINTDAQGFNKKVSSAASNLQKQLPGLKIVIFDIYKPLYDLVQNPSNF 311
>TC90972 similar to GP|10638955|emb|CAB81548. putative proline-rich protein
APG isolog {Cicer arietinum}, partial (88%)
Length = 1258
Score = 454 bits (1167), Expect = e-128
Identities = 217/289 (75%), Positives = 252/289 (87%)
Frame = +3
Query: 2 MNMNSKETLVLLIVSCFLTCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYG 61
M MNS+E ++ FL G+ DT+VPAI+TFGDSAVDVGNNDYL TLFKANYPPYG
Sbjct: 18 MKMNSREAFFVVFAFVFLGWGNAQDDTVVPAIVTFGDSAVDVGNNDYLFTLFKANYPPYG 197
Query: 62 RDFTNKQPTGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASGYDE 121
RDF +PTGRFCNGKLATD TAETLGF S+APAYLSPQA+GKNLL+GANFASAASGYDE
Sbjct: 198 RDFVXHKPTGRFCNGKLATDITAETLGFKSYAPAYLSPQATGKNLLIGANFASAASGYDE 377
Query: 122 KAATLNHAIPLSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTNP 181
KAA LNHAIPLSQQL+Y+KEYQ KL+++AGSKKAASIIK +LY+LS GSSDF+QNYY NP
Sbjct: 378 KAAILNHAIPLSQQLKYYKEYQSKLSKIAGSKKAASIIKGALYLLSGGSSDFIQNYYVNP 557
Query: 182 WINQAITVDQYSSYLLDSFTNFIKGVYGLGARKIGVTSLPPLGCLPAARTLFGYHENGCV 241
IN+ +T DQYS+YL+D++++F+K +Y LGARKIGVTSLPPLGCLPA RTLFG+HE GCV
Sbjct: 558 LINKVVTPDQYSAYLVDTYSSFVKDLYKLGARKIGVTSLPPLGCLPATRTLFGFHEKGCV 737
Query: 242 ARINTDAQGFNKKVSSAASNLQKQLPGLKIVIFDIYKPLYDLVQNPSNF 290
RIN DAQGFNKK++SA LQKQLPGLKIV+F+IYKPLY+LVQ+PS F
Sbjct: 738 TRINNDAQGFNKKINSATVKLQKQLPGLKIVVFNIYKPLYELVQSPSKF 884
>TC80740 similar to GP|10177228|dbj|BAB10602. GDSL-motif
lipase/hydrolase-like protein {Arabidopsis thaliana},
partial (94%)
Length = 1167
Score = 320 bits (821), Expect = 3e-88
Identities = 156/260 (60%), Positives = 192/260 (73%)
Frame = +3
Query: 29 LVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQPTGRFCNGKLATDFTAETLG 88
LVPA+ FGDS VD NN+ L T+ K+N+PPYGRDF N+ PTGRFCNGKLA DFTAE LG
Sbjct: 180 LVPALFIFGDSVVDARNNNNLYTIVKSNFPPYGRDFNNQMPTGRFCNGKLAADFTAENLG 359
Query: 89 FTSFAPAYLSPQASGKNLLLGANFASAASGYDEKAATLNHAIPLSQQLEYFKEYQGKLAQ 148
FT++ PAYL+ Q KNLL GANFAS ASGY + A L HAI L QQLE++KE Q L
Sbjct: 360 FTTYPPAYLNLQEKRKNLLNGANFASGASGYFDPTAKLYHAISLEQQLEHYKECQNILVG 539
Query: 149 VAGSKKAASIIKDSLYVLSAGSSDFVQNYYTNPWINQAITVDQYSSYLLDSFTNFIKGVY 208
VAG A+SII ++Y++ AGSSDFVQNYY NP + + T DQ+S L+ +T FI+ +Y
Sbjct: 540 VAGKSNASSIISGAIYLVRAGSSDFVQNYYINPLLYKVFTADQFSDILMQHYTIFIQNLY 719
Query: 209 GLGARKIGVTSLPPLGCLPAARTLFGYHENGCVARINTDAQGFNKKVSSAASNLQKQLPG 268
LGARKIGVT+LPPLGCLPAA TLFG H N CV R+N DA FN K+++ + NLQK+L
Sbjct: 720 ALGARKIGVTTLPPLGCLPAAITLFGSHSNECVDRLNNDALNFNTKLNTTSQNLQKELSN 899
Query: 269 LKIVIFDIYKPLYDLVQNPS 288
L + + DIY+PL+DLV P+
Sbjct: 900 LTLAVLDIYQPLHDLVTKPT 959
>TC80033 weakly similar to GP|21593518|gb|AAM65485.1 putative GDSL-motif
lipase/hydrolase {Arabidopsis thaliana}, partial (89%)
Length = 1164
Score = 238 bits (606), Expect = 3e-63
Identities = 117/278 (42%), Positives = 170/278 (61%)
Frame = +1
Query: 10 LVLLIVSCFLTCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQP 69
L L + C L VPAI+ FGDS+VD GNN+++PT+ ++N+ PYGRDF +
Sbjct: 67 LCSLHILCLLLFHLNKVSAKVPAIIVFGDSSVDAGNNNFIPTVARSNFQPYGRDFQGGKA 246
Query: 70 TGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASGYDEKAATLNHA 129
TGRF NG++ TDF AE+ G PAYL P+ + + G +FASAA+GYD + +
Sbjct: 247 TGRFSNGRIPTDFIAESFGIKESVPAYLDPKYNISDFATGVSFASAATGYDNATSDVLSV 426
Query: 130 IPLSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTNPWINQAITV 189
IPL +QLEY+K+YQ L+ G KA I +S++++S G++DF++NYYT P T
Sbjct: 427 IPLWKQLEYYKDYQKNLSSYLGEAKAKETISESVHLMSMGTNDFLENYYTMPGRASQYTP 606
Query: 190 DQYSSYLLDSFTNFIKGVYGLGARKIGVTSLPPLGCLPAARTLFGYHENGCVARINTDAQ 249
QY ++L NFI+ +Y LGARKI + LPP+GCLP RT +NGCVA N A
Sbjct: 607 QQYQTFLAGIAENFIRNLYALGARKISLGGLPPMGCLPLERTTNFMGQNGCVANFNNIAL 786
Query: 250 GFNKKVSSAASNLQKQLPGLKIVIFDIYKPLYDLVQNP 287
FN K+ + + L ++LP +K+V + Y + +++ P
Sbjct: 787 EFNDKLKNITTKLNQELPDMKLVFSNPYYIMLHIIKKP 900
>BG451611 homologue to GP|10638955|em putative proline-rich protein APG
isolog {Cicer arietinum}, partial (32%)
Length = 645
Score = 203 bits (516), Expect(2) = 3e-61
Identities = 98/98 (100%), Positives = 98/98 (100%)
Frame = +3
Query: 30 VPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQPTGRFCNGKLATDFTAETLGF 89
VPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQPTGRFCNGKLATDFTAETLGF
Sbjct: 96 VPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQPTGRFCNGKLATDFTAETLGF 275
Query: 90 TSFAPAYLSPQASGKNLLLGANFASAASGYDEKAATLN 127
TSFAPAYLSPQASGKNLLLGANFASAASGYDEKAATLN
Sbjct: 276 TSFAPAYLSPQASGKNLLLGANFASAASGYDEKAATLN 389
Score = 49.7 bits (117), Expect(2) = 3e-61
Identities = 23/23 (100%), Positives = 23/23 (100%)
Frame = +1
Query: 1 MMNMNSKETLVLLIVSCFLTCGS 23
MMNMNSKETLVLLIVSCFLTCGS
Sbjct: 7 MMNMNSKETLVLLIVSCFLTCGS 75
>TC91569 similar to GP|18252233|gb|AAL61949.1 putative APG protein
{Arabidopsis thaliana}, partial (60%)
Length = 1398
Score = 224 bits (572), Expect = 2e-59
Identities = 113/282 (40%), Positives = 169/282 (59%), Gaps = 3/282 (1%)
Frame = +1
Query: 10 LVLLIVSCFLTCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQP 69
L+L+ +TC + + VPA++ FGDS+VD GNN+ + T K+N+ PYGRD +P
Sbjct: 76 LILITQMIMVTCNN---ENYVPAVIVFGDSSVDSGNNNMISTFLKSNFRPYGRDIDGGRP 246
Query: 70 TGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASGYDEKAATLNHA 129
TGRF NG++ DF +E G S PAYL P + + + G FASA +GYD + + +
Sbjct: 247 TGRFSNGRIPPDFISEAFGIKSLIPAYLDPAYTIDDFVTGVCFASAGTGYDNATSAILNV 426
Query: 130 IPLSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTNPWINQAITV 189
IPL +++E++KEYQ KL G +K+ II ++LY++S G++DF+ NYY + T+
Sbjct: 427 IPLWKEVEFYKEYQDKLKAHIGEEKSIEIISEALYIISLGTNDFLGNYYGFTTLRFRYTI 606
Query: 190 DQYSSYLLDSFTNFIKGVYGLGARKIGVTSLPPLGCLPAARTL---FGYHENGCVARINT 246
QY YL+ NFI+ +Y LGARK+ +T L P+GCLP R + G+H C + N
Sbjct: 607 SQYQDYLIGIAENFIRQLYSLGARKLAITGLIPMGCLPLERAINIFGGFHR--CYEKYNI 780
Query: 247 DAQGFNKKVSSAASNLQKQLPGLKIVIFDIYKPLYDLVQNPS 288
A FN K+ + S L K+LP LK + ++Y D++ PS
Sbjct: 781 VALEFNVKLENMISKLNKELPQLKALSANVYDLFNDIITRPS 906
>TC80992 weakly similar to GP|21593518|gb|AAM65485.1 putative GDSL-motif
lipase/hydrolase {Arabidopsis thaliana}, partial (53%)
Length = 812
Score = 186 bits (473), Expect = 7e-48
Identities = 92/209 (44%), Positives = 133/209 (63%)
Frame = +1
Query: 10 LVLLIVSCFLTCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQP 69
L+L+ +TC + VPA++ FGDS+VD GNN+ + TL K+N+ PYGRDF +P
Sbjct: 82 LILITQIIMVTCKT---KNHVPAVIVFGDSSVDSGNNNRIATLLKSNFKPYGRDFEGGRP 252
Query: 70 TGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASGYDEKAATLNHA 129
TGRFCNG+ DF AE G PAYL P + + + G FASA +GYD + + +
Sbjct: 253 TGRFCNGRTPPDFIAEAFGVKRNIPAYLDPAYTIDDFVTGVCFASAGTGYDNATSDVLNV 432
Query: 130 IPLSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTNPWINQAITV 189
IPL +++E+FKEYQ KL G KKA II ++LY++S G++DF++NYY P TV
Sbjct: 433 IPLWKEIEFFKEYQEKLRVHVGKKKANEIISEALYLISLGTNDFLENYYIFPTRQLHFTV 612
Query: 190 DQYSSYLLDSFTNFIKGVYGLGARKIGVT 218
QY +L+D +F++ ++ LGARK+ +T
Sbjct: 613 SQYQDFLVDIAEDFVRKLHSLGARKLSIT 699
>CA990150 weakly similar to GP|10177228|db GDSL-motif lipase/hydrolase-like
protein {Arabidopsis thaliana}, partial (35%)
Length = 874
Score = 176 bits (447), Expect = 7e-45
Identities = 83/163 (50%), Positives = 113/163 (68%)
Frame = +1
Query: 128 HAIPLSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTNPWINQAI 187
H IPL QLE++K+ Q +L ++ G + A SII + Y+L GS DF QNY+ NP +
Sbjct: 247 HVIPLMNQLEFYKDCQEELVKLVGKENATSIISGAAYLLVDGSGDFAQNYFINPILQNLY 426
Query: 188 TVDQYSSYLLDSFTNFIKGVYGLGARKIGVTSLPPLGCLPAARTLFGYHENGCVARINTD 247
T Q+S L++ + NFI+ +Y LGARKIGVT+LPP+GC+P T FGYH N CV IN
Sbjct: 427 TPYQFSDVLIEEYYNFIQNLYALGARKIGVTTLPPIGCMPFIITKFGYHSNKCVETINNV 606
Query: 248 AQGFNKKVSSAASNLQKQLPGLKIVIFDIYKPLYDLVQNPSNF 290
A FNKK++ NL K+LPG+K+VIFDIY+PLY+L+ PS++
Sbjct: 607 AIYFNKKLNLTTENLIKKLPGVKLVIFDIYQPLYELIIRPSDY 735
Score = 38.1 bits (87), Expect = 0.004
Identities = 16/33 (48%), Positives = 22/33 (66%)
Frame = +2
Query: 92 FAPAYLSPQASGKNLLLGANFASAASGYDEKAA 124
+ PAYL+ G N+L GANFAS+ SGY + +
Sbjct: 2 YQPAYLNLNTKGSNILNGANFASSGSGYHDSTS 100
>TC89337 similar to GP|21592967|gb|AAM64916.1 putative GDSL-motif
lipase/acylhydrolase {Arabidopsis thaliana}, partial
(65%)
Length = 881
Score = 157 bits (396), Expect = 6e-39
Identities = 95/273 (34%), Positives = 153/273 (55%), Gaps = 7/273 (2%)
Frame = +1
Query: 1 MMNMNSKETLVLLIVSCFLTCGSFAQD--TLVPAIMTFGDSAVDVGNNDYLPTLFKANYP 58
M +++S LV+L+V G F + + + FGDS VD GNN+YL T +A+ P
Sbjct: 34 MASLSSFVALVILVVG-----GIFVHEIEAIPRTFLVFGDSLVDNGNNNYLATTARADAP 198
Query: 59 PYGRDFT-NKQPTGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAAS 117
PYG D+ + +PTGRF NG D ++ LG P YLSP+ G+ LL+GANFASA
Sbjct: 199 PYGIDYQPSHRPTGRFSNGYNIPDIISQKLGAEPTLP-YLSPELRGEKLLVGANFASAGI 375
Query: 118 GY-DEKAATLNHAIPLSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQN 176
G ++ + I + +Q EYF+EYQ +L+ + G+ +A S + +L +++ G +DFV N
Sbjct: 376 GILNDTGIQFINIIRMYRQYEYFQEYQSRLSALIGASQAKSRVNQALVLITVGGNDFVNN 555
Query: 177 YYTNPWI--NQAITVDQYSSYLLDSFTNFIKGVYGLGARKIGVTSLPPLGCLPAARTLFG 234
YY P+ ++ + +Y YL+ + ++ +Y LGAR++ VT P+GC+P+ G
Sbjct: 556 YYLVPYSARSRQYPLPEYVKYLISEYQKLLQKLYDLGARRVLVTGTGPMGCVPSEIAQRG 735
Query: 235 YHENG-CVARINTDAQGFNKKVSSAASNLQKQL 266
NG C + + FN ++ + L K++
Sbjct: 736 --RNGQCSTELQRASSLFNPQLENMLLGLNKKI 828
>TC88477 similar to GP|21592417|gb|AAM64368.1 lipase/hydrolase putative
{Arabidopsis thaliana}, partial (92%)
Length = 1415
Score = 153 bits (387), Expect = 7e-38
Identities = 99/283 (34%), Positives = 147/283 (50%), Gaps = 4/283 (1%)
Frame = +2
Query: 12 LLIVSCFLTCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQPTG 71
LL+V L G A D VP FGDS VD GNN+ L +L +A+Y PYG DF PTG
Sbjct: 125 LLVVVLGLWSGVGA-DPQVPWYFIFGDSLVDNGNNNGLQSLARADYLPYGIDFGG--PTG 295
Query: 72 RFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASGY-DEKAATLNHAI 130
RF NGK D AE LGF + P Y S AS +L G N+ASAA+G +E L +
Sbjct: 296 RFSNGKTTVDAIAELLGFDDYIPPYAS--ASDDAILKGVNYASAAAGIREETGRQLGARL 469
Query: 131 PLSQQLEYFKEYQGKLAQVAGSK-KAASIIKDSLYVLSAGSSDFVQNYYTNPWIN--QAI 187
S Q++ ++ ++ + G++ +AAS + +Y + GS+D++ NY+ + N
Sbjct: 470 SFSAQVQNYQSTVSQVVNILGTEDQAASHLSKCIYSIGLGSNDYLNNYFMPQFYNTHDQY 649
Query: 188 TVDQYSSYLLDSFTNFIKGVYGLGARKIGVTSLPPLGCLPAARTLFGYHENGCVARINTD 247
T D+Y+ L+ S+T ++ +Y ARK+ + + +GC P CV IN+
Sbjct: 650 TPDEYADDLIQSYTEQLRTLYNN*ARKMVLFGIGQIGCSPNELATRSADGVTCVEEINSA 829
Query: 248 AQGFNKKVSSAASNLQKQLPGLKIVIFDIYKPLYDLVQNPSNF 290
Q FN K+ QLP K++ + Y D++ NPS +
Sbjct: 830 NQIFNNKLKGLVDQFNNQLPDSKVIYVNSYGIFQDIISNPSAY 958
>TC83268 weakly similar to GP|21592974|gb|AAM64923.1 proline-rich protein
putative {Arabidopsis thaliana}, partial (40%)
Length = 1156
Score = 152 bits (384), Expect = 1e-37
Identities = 79/212 (37%), Positives = 118/212 (55%), Gaps = 1/212 (0%)
Frame = +3
Query: 77 KLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASGYDEKAATLNHAIPLSQQL 136
KLA DF A TL P +L P S + LL G +FAS SG+D+ L AI +S+Q+
Sbjct: 3 KLAIDFLASTLNLKETVPPFLDPNLSNEELLKGVSFASGGSGFDDFTIALTGAISMSKQV 182
Query: 137 EYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTNPWINQAITVDQYSSYL 196
EYFK+Y K+ + G K+A + ++L ++SAG++DF+ N+Y P + Y Y+
Sbjct: 183 EYFKDYVHKVKSIVGEKEAKQRVGNALVIISAGTNDFLFNFYDIPTRRLEFNISGYQDYV 362
Query: 197 LDSFTNFIKGVYGLGARKIGVTSLPPLGCLPAARTL-FGYHENGCVARINTDAQGFNKKV 255
FIK +Y LG RK V LPP+GC+P T F CV N +A+ +N+K+
Sbjct: 363 QSRLLIFIKELYELGCRKFAVAGLPPIGCIPVQITAKFVKDRYKCVKEENLEAKDYNQKL 542
Query: 256 SSAASNLQKQLPGLKIVIFDIYKPLYDLVQNP 287
+ LQ L G +++ +IY PL L+++P
Sbjct: 543 ARRLLQLQAILSGSRVIYTNIYDPLIGLIKHP 638
>BI273040 similar to GP|6721157|gb| putative GDSL-motif lipase/acylhydrolase
{Arabidopsis thaliana}, partial (60%)
Length = 692
Score = 149 bits (377), Expect = 9e-37
Identities = 83/227 (36%), Positives = 135/227 (58%), Gaps = 9/227 (3%)
Frame = +2
Query: 27 DTLVP-----AIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQPTGRFCNGKLATD 81
+T+VP A FGDS VD GNN+YL T +A+ PYG D+ + TGRF NG D
Sbjct: 17 NTVVPQVEARAFFVFGDSLVDNGNNNYLATTARADSYPYGIDYPTHRATGRFSNGLNMPD 196
Query: 82 FTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASG-YDEKAATLNHAIPLSQQLEYFK 140
+E +G P YLSP+ +G+ LL+GANFASA G ++ + I +++QL+YF+
Sbjct: 197 LISERIGSQPTLP-YLSPELNGEALLVGANFASAGIGILNDTGIQFFNIIRITRQLQYFE 373
Query: 141 EYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTNPWI--NQAITVDQYSSYLLD 198
+YQ +++ + G ++ ++ ++LY+++ G +DFV NY+ P+ ++ + Y YL+
Sbjct: 374 QYQQRVSALIGEEETVRLVNEALYLMTLGGNDFVNNYFLVPFSARSRQFXLPDYVVYLIS 553
Query: 199 SFTNFIKGVYGLGARKIGVTSLPPLGCLPAARTLFGYHENG-CVARI 244
+ + +Y LGAR++ VT PLGC+P L + +NG C A++
Sbjct: 554 EYRKILARLYELGARRVLVTGTXPLGCVPX--XLXXHXKNGECYAKL 688
>BE319090 weakly similar to GP|15054386|gb family II lipase EXL3 {Arabidopsis
thaliana}, partial (42%)
Length = 664
Score = 149 bits (377), Expect = 9e-37
Identities = 76/184 (41%), Positives = 101/184 (54%)
Frame = +1
Query: 31 PAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQPTGRFCNGKLATDFTAETLGFT 90
PA+ FGDS +D GNN+ PT + +PPYG+DF PTGRF NGK+ D E LG
Sbjct: 112 PAVFVFGDSIMDTGNNNNRPTPTQCKFPPYGKDFQGGIPTGRFSNGKVPADLIVEELGIK 291
Query: 91 SFAPAYLSPQASGKNLLLGANFASAASGYDEKAATLNHAIPLSQQLEYFKEYQGKLAQVA 150
+ PAYL P L+ G NFAS +GYD + + AI +S Q+E FKEY KL +
Sbjct: 292 EYLPAYLDPNLQPSELVTGVNFASGGAGYDPLTSKIEAAISMSAQIELFKEYIVKLKGIV 471
Query: 151 GSKKAASIIKDSLYVLSAGSSDFVQNYYTNPWINQAITVDQYSSYLLDSFTNFIKGVYGL 210
G + I+ +S+Y + GS+D Y+ YS L+DS NF K +Y L
Sbjct: 472 GEDRTNFILANSIYFVLVGSNDISNTYFLFHARQVNYDFPSYSDLLVDSAYNFYKEMYQL 651
Query: 211 GARK 214
GAR+
Sbjct: 652 GARR 663
>TC91750 similar to GP|6721157|gb|AAF26785.1| putative GDSL-motif
lipase/acylhydrolase {Arabidopsis thaliana}, partial
(50%)
Length = 686
Score = 143 bits (361), Expect = 7e-35
Identities = 79/208 (37%), Positives = 126/208 (59%), Gaps = 3/208 (1%)
Frame = +3
Query: 10 LVLLIVSCFLTCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQP 69
L++ + SCF G+ AQ A FGDS VD GNN+YL T +A+ PPYG D+ ++P
Sbjct: 66 LMVALTSCFK--GTVAQR----AFFVFGDSLVDNGNNNYLATTARADAPPYGIDYPTRRP 227
Query: 70 TGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASG-YDEKAATLNH 128
TGRF NG DF ++ LG P YLSP+ +G+ LL+GANFASA G ++ +
Sbjct: 228 TGRFSNGYNIPDFISQALGAEPTLP-YLSPELNGEALLVGANFASAGIGILNDTGIQFIN 404
Query: 129 AIPLSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTNPWI--NQA 186
I + +QLEYF++YQ +++ + G ++ S++ +L +++ G +DFV NYY P+ ++
Sbjct: 405 IIRIFRQLEYFQQYQQRVSGLIGPEQTQSLVNGALVLITLGGNDFVNNYYLVPFSARSRQ 584
Query: 187 ITVDQYSSYLLDSFTNFIKGVYGLGARK 214
+ Y Y++ + ++ +Y LGAR+
Sbjct: 585 YNLPDYVRYIISEYKKILRRLYHLGARR 668
>TC88213 weakly similar to GP|15054386|gb|AAK30018.1 family II lipase EXL3
{Arabidopsis thaliana}, partial (43%)
Length = 677
Score = 139 bits (350), Expect = 1e-33
Identities = 67/150 (44%), Positives = 89/150 (58%)
Frame = +1
Query: 30 VPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQPTGRFCNGKLATDFTAETLGF 89
+PA++ FGDS +D GNN+ + T+ K N+PPYG+DF PTGRFCNGK +D E LG
Sbjct: 181 IPALIAFGDSIMDTGNNNNIKTIVKCNFPPYGQDFEGGIPTGRFCNGKNPSDLIVEELGI 360
Query: 90 TSFAPAYLSPQASGKNLLLGANFASAASGYDEKAATLNHAIPLSQQLEYFKEYQGKLAQV 149
PAYL P +L G FAS ASGYD + I + QL+ FKEY KL V
Sbjct: 361 KELLPAYLDPNLKPSDLSTGVCFASGASGYDPLTPKIVSVISMGDQLKMFKEYIVKLKGV 540
Query: 150 AGSKKAASIIKDSLYVLSAGSSDFVQNYYT 179
G +A I+ ++L+++ AGS D Y+T
Sbjct: 541 VGENRANFILTNTLFLIVAGSDDLANTYFT 630
>TC80438 similar to PIR|E96579|E96579 hypothetical protein T18A20.15
[imported] - Arabidopsis thaliana, partial (28%)
Length = 857
Score = 139 bits (350), Expect = 1e-33
Identities = 87/240 (36%), Positives = 127/240 (52%), Gaps = 3/240 (1%)
Frame = +2
Query: 32 AIMTFGDSAVDVGNNDYLPTL--FKANYPPYGRDFTNKQPTGRFCNGKLATDFTAETLGF 89
A+ FGDS VD GNN+Y+ T+ KA+ PYG++ +PTGRF +G++ TDF AE
Sbjct: 143 ALFIFGDSTVDSGNNNYIDTIPENKADCKPYGQNGVFDKPTGRFSDGRVITDFIAEYAKL 322
Query: 90 TSFAPAYLSPQASGKNLLLGANFASAASGYDEKAATLNHAIPLSQQLEYFKEYQGKLAQV 149
P YL P N G NFAS +G + I L QL F+E + LA+
Sbjct: 323 P-LIPPYLKPSIDYSN---GVNFASGGAGVLPET-NQGLVIDLPTQLSNFEEVRKSLAEK 487
Query: 150 AGSKKAASIIKDSLYVLSAGSSDFVQNYYTNPWINQAITVDQYSSYLLDSFTNFIKGVYG 209
G +KA +I +++Y +S GS+D++ Y NP + ++ QY ++ + T I +Y
Sbjct: 488 LGEEKAKELISEAVYFISIGSNDYMGGYLGNPKMQESYNPQQYIGMVIGNLTQSIVRLYE 667
Query: 210 LGARKIGVTSLPPLGCLPAARTLF-GYHENGCVARINTDAQGFNKKVSSAASNLQKQLPG 268
GARK G SL PLGCLPA R + GC ++ A N +S+ ++L + L G
Sbjct: 668 KGARKFGFLSLSPLGCLPALRAANPEASKGGCFGAASSLALAHNNALSNILTSLNQVLKG 847
>TC86678 similar to PIR|E96579|E96579 hypothetical protein T18A20.15
[imported] - Arabidopsis thaliana, partial (43%)
Length = 1341
Score = 139 bits (350), Expect = 1e-33
Identities = 82/261 (31%), Positives = 137/261 (52%), Gaps = 2/261 (0%)
Frame = +2
Query: 32 AIMTFGDSAVDVGNNDYL--PTLFKANYPPYGRDFTNKQPTGRFCNGKLATDFTAETLGF 89
A+ FGDS +D GNN+Y+ T +AN+ PYG + N PTGRF +G+L +DF AE +
Sbjct: 182 ALFIFGDSFLDAGNNNYINTTTFDQANFLPYGETYFNF-PTGRFSDGRLISDFIAEYVNI 358
Query: 90 TSFAPAYLSPQASGKNLLLGANFASAASGYDEKAATLNHAIPLSQQLEYFKEYQGKLAQV 149
P +L P G NFAS +G + IP Q FK+ L
Sbjct: 359 P-LVPPFLQPD--NNKYYNGVNFASGGAGALVETFQ-GSVIPFKTQAINFKKVTTWLRHK 526
Query: 150 AGSKKAASIIKDSLYVLSAGSSDFVQNYYTNPWINQAITVDQYSSYLLDSFTNFIKGVYG 209
GS + +++ +++Y+ S GS+D++ + TN + + + +Y + ++ +FT+ IK ++
Sbjct: 527 LGSSDSKTLLSNAVYMFSIGSNDYLSPFLTNSDVLKHYSHTEYVAMVIGNFTSTIKEIHK 706
Query: 210 LGARKIGVTSLPPLGCLPAARTLFGYHENGCVARINTDAQGFNKKVSSAASNLQKQLPGL 269
GA+K + +LPPLGCLP R + + C+ +++ A N+ + LQKQL G
Sbjct: 707 RGAKKFVILNLPPLGCLPGTRIIQSQGKGSCLEELSSLASIHNQALYEVLLELQKQLRGF 886
Query: 270 KIVIFDIYKPLYDLVQNPSNF 290
K ++D L ++ +P +
Sbjct: 887 KFSLYDFNSDLSHMINHPLKY 949
>TC79259 similar to GP|6721157|gb|AAF26785.1| putative GDSL-motif
lipase/acylhydrolase {Arabidopsis thaliana}, partial
(52%)
Length = 712
Score = 130 bits (326), Expect = 8e-31
Identities = 78/218 (35%), Positives = 123/218 (55%), Gaps = 3/218 (1%)
Frame = +3
Query: 4 MNSKETLVLLIVSCFLTCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRD 63
M + + ++ F+ C ++AQ A FGDS VD GNND+L T +A+ PYG D
Sbjct: 66 MATSLVIAFCVMISFVGC-AYAQPR---AFGVFGDSLVDSGNNDFLATTARADNYPYGID 233
Query: 64 FTNKQPTGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASG-YDEK 122
+ + +PTGRF NG D + LG P YLSP G+ LL+GANFASA G ++
Sbjct: 234 YPSHRPTGRFSNGYNIPDLISLELGLEPTLP-YLSPLLVGEKLLIGANFASAGIGILNDT 410
Query: 123 AATLNHAIPLSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTNPW 182
H I + +QL F+ YQ +++ GS+ A +++ +L +++ G +DFV NYY P+
Sbjct: 411 GFQFIHIIRIYKQLRLFELYQKRVSAHIGSEGARNLVNRALVLITLGGNDFVNNYYLVPF 590
Query: 183 I--NQAITVDQYSSYLLDSFTNFIKGVYGLGARKIGVT 218
++ ++ Y YL+ + ++ +Y LGAR++ VT
Sbjct: 591 SARSRQFSLPDYVRYLISEYRKVLRRLYDLGARRVLVT 704
>TC87519 weakly similar to GP|21592967|gb|AAM64916.1 putative GDSL-motif
lipase/acylhydrolase {Arabidopsis thaliana}, partial
(76%)
Length = 1084
Score = 127 bits (318), Expect = 7e-30
Identities = 71/207 (34%), Positives = 116/207 (55%), Gaps = 3/207 (1%)
Frame = +2
Query: 63 DFTNKQPTGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASGY-DE 121
DF +QPTGRF NG D ++ LG + P YLSP+ G +L GANFASA G ++
Sbjct: 2 DFPTRQPTGRFSNGLNVPDLISKELGSSPPLP-YLSPKLRGHRMLNGANFASAGIGILND 178
Query: 122 KAATLNHAIPLSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQNYY--T 179
I + +QL++F+EYQ +++ + G K+A +I +L +++ G +DFV NYY
Sbjct: 179 TGFQFIEVIRMYKQLDFFEEYQKRVSDLIGKKEAKKLINGALILITCGGNDFVNNYYLVP 358
Query: 180 NPWINQAITVDQYSSYLLDSFTNFIKGVYGLGARKIGVTSLPPLGCLPAARTLFGYHENG 239
N ++ + +Y +YLL + ++ +Y LGAR++ V+ P+GC PAA + G +
Sbjct: 359 NSLRSRQYALPEYVTYLLSEYKKILRRLYHLGARRVLVSGTGPMGCAPAALAI-GGTDGE 535
Query: 240 CVARINTDAQGFNKKVSSAASNLQKQL 266
C + A +N K+ + L +Q+
Sbjct: 536 CAPELQLAASLYNPKLVQLITELNQQI 616
>TC83569 weakly similar to GP|6733842|emb|CAB69346.1 unnamed protein product
{unidentified}, partial (39%)
Length = 1185
Score = 118 bits (296), Expect = 2e-27
Identities = 78/224 (34%), Positives = 120/224 (52%), Gaps = 5/224 (2%)
Frame = +1
Query: 45 NNDYLPTLFKANYPPYGRDFTNKQPTGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGK 104
NN+ LPT K+NY PYG DF PTGRF NG+ + D + LGF F P + + +G
Sbjct: 1 NNNNLPTSAKSNYKPYGIDFP-MGPTGRFTNGRTSIDIITQLLGFEKFIPPFAN--INGS 171
Query: 105 NLLLGANFASAASGYD-EKAATLNHAIPLSQQLEYFKEYQGKLA-QVAGSKKAASIIKDS 162
++L G N+AS A+G E + T I L QLE K ++A ++ G KA +
Sbjct: 172 DILKGVNYASGAAGIRIETSITTGFVISLGLQLENHKVIVSRIASRLEGIDKAQEYLSKC 351
Query: 163 LYVLSAGSSDFVQNYYTNPW--INQAITVDQYSSYLLDSFTNFIKGVYGLGARKIGVTSL 220
LY ++ GS+D++ NY+ + +Q + +QY+ L+ + + ++ +GARK + L
Sbjct: 352 LYYVNIGSNDYINNYFRPQFYPTSQIYSPEQYAEALIQELSLNLLTLHDIGARKYVLVGL 531
Query: 221 PPLGCLPAARTLFGYHENG-CVARINTDAQGFNKKVSSAASNLQ 263
LGC P+A +F + NG CV N A FN K+ + + Q
Sbjct: 532 GLLGCTPSA--IFTHGTNGSCVDEENAPALIFNFKLKFFSGSFQ 657
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.135 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,056,068
Number of Sequences: 36976
Number of extensions: 96308
Number of successful extensions: 595
Number of sequences better than 10.0: 95
Number of HSP's better than 10.0 without gapping: 524
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 526
length of query: 291
length of database: 9,014,727
effective HSP length: 95
effective length of query: 196
effective length of database: 5,502,007
effective search space: 1078393372
effective search space used: 1078393372
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146586.11


