

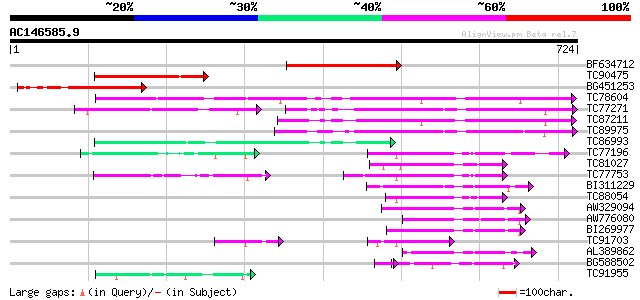
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146585.9 - phase: 0
(724 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BF634712 similar to PIR|T07943|T07 probable AMP-binding protein ... 229 4e-60
TC90475 similar to GP|17065456|gb|AAL32882.1 A6 anther-specific ... 218 6e-57
BG451253 similar to GP|17065456|gb| A6 anther-specific protein {... 164 1e-40
TC78604 similar to PIR|E84456|E84456 probable acyl-CoA synthetas... 153 3e-37
TC77271 similar to PIR|T07929|T07929 probable long-chain-fatty-a... 98 1e-36
TC87211 similar to GP|13516481|dbj|BAB40450. AMP-binding protein... 129 4e-30
TC89975 similar to PIR|G96530|G96530 probable acyl CoA synthetas... 104 1e-22
TC86993 similar to GP|13516481|dbj|BAB40450. AMP-binding protein... 96 5e-20
TC77196 similar to GP|17063848|gb|AAL35216.1 4-coumarate:CoA lig... 90 3e-18
TC81027 similar to PIR|H85064|H85064 4-coumarate--CoA ligase-lik... 89 8e-18
TC77753 similar to GP|19773582|gb|AAL98709.1 4-coumarate:coenzym... 86 7e-17
BI311229 similar to GP|20502991|gb Putative AMP-binding protein ... 85 1e-16
TC88054 homologue to PIR|PQ0772|PQ0772 4-coumarate--CoA ligase (... 83 4e-16
AW329094 similar to GP|20502991|gb Putative AMP-binding protein ... 81 1e-15
AW776080 similar to GP|20161027|db putative 4-coumarate-CoA liga... 79 6e-15
BI269977 similar to GP|12039389|gb putative 4-coumarate CoA liga... 79 8e-15
TC91703 similar to GP|4038975|gb|AAC97600.1| 4-coumarate:CoA lig... 75 7e-14
AL389862 similar to GP|23315116|gb Sequence 281 from patent US 6... 71 2e-12
BG588502 SP|P31552|CAIC Probable crotonobetaine/carnitine-CoA li... 58 2e-11
TC91955 weakly similar to PIR|T27421|T27421 hypothetical protein... 62 8e-10
>BF634712 similar to PIR|T07943|T07 probable AMP-binding protein - rape,
partial (20%)
Length = 668
Score = 229 bits (583), Expect = 4e-60
Identities = 119/147 (80%), Positives = 129/147 (86%)
Frame = +2
Query: 354 MISVPLVFETLYSGIQKQISTSPPVRKLVALTFIRVSLAYMEYKRIYEGKCLTRNVKQPS 413
+ISVPLV+E+LYSGIQ+QISTS VRKLVALTFIRVSL YME KRIYEGKCLT+N K PS
Sbjct: 2 LISVPLVYESLYSGIQRQISTSSLVRKLVALTFIRVSLGYMECKRIYEGKCLTKNQKSPS 181
Query: 414 IVNSMLDCLWARIIATILFPIHLLAIKFVYSKIHSAIGLSKAGISGGGSLPLEVDKFFEA 473
+ +MLD L ARIIATILFPIH+LA K VYSKIHSAIG SKAGISGGGSLP VD+FFEA
Sbjct: 182 YLYAMLDWLGARIIATILFPIHMLAKKLVYSKIHSAIGFSKAGISGGGSLPSHVDRFFEA 361
Query: 474 IGVKVQNGYGLTETSPVIAARRPRCNV 500
IGV +QNGYGLTETSPVIAARR CNV
Sbjct: 362 IGVTLQNGYGLTETSPVIAARRLSCNV 442
>TC90475 similar to GP|17065456|gb|AAL32882.1 A6 anther-specific protein
{Arabidopsis thaliana}, partial (15%)
Length = 761
Score = 218 bits (555), Expect = 6e-57
Identities = 111/146 (76%), Positives = 129/146 (88%)
Frame = +2
Query: 109 ITYNQLEQAILDYAEGLRVIGVRPDEKLALFADNSCRWLVADQGMMASGAINVVRGSRSS 168
+T QL+ AIL++AEGLRVIGV+PDEK+ALFADNSCRWLVADQGMMA GAINVVRGSRSS
Sbjct: 185 LTTFQLKDAILNFAEGLRVIGVKPDEKIALFADNSCRWLVADQGMMAIGAINVVRGSRSS 364
Query: 169 VEELLQIYNHSESVALAVDGPEMFNRIAKPFYSKTGMRFIILLWGEKSDLNLIAEENKEV 228
+EELLQIYNHSESVALAVD PEM+N+IAKPFY KT +RFIILLWGEKS +L+ E +K V
Sbjct: 365 IEELLQIYNHSESVALAVDNPEMYNQIAKPFYLKTSIRFIILLWGEKS--SLVNEADKGV 538
Query: 229 PIFSFMEVIDLGRESRMALSDSHEAS 254
PI++FMEVI+ GR+SR AL S +AS
Sbjct: 539 PIYTFMEVINFGRQSRRALHTSDDAS 616
>BG451253 similar to GP|17065456|gb| A6 anther-specific protein {Arabidopsis
thaliana}, partial (12%)
Length = 686
Score = 164 bits (414), Expect = 1e-40
Identities = 96/166 (57%), Positives = 113/166 (67%), Gaps = 2/166 (1%)
Frame = +1
Query: 11 FSYHFFTTTTITLPRTNYARPRCFRVFSQSNTETVQIIRKCSPFLESSMLVGNNNNNNNA 70
F H FTT+ I L RT R+F QS ++ IR+ SP LESS+L GN+
Sbjct: 205 FPRHSFTTS-INLSRT--------RIFCQSQSK----IRRFSPLLESSLLSGNDGV---- 333
Query: 71 AVDSSHEWKVVPDIWRSSAEKYGDKVALVDQYHHPPSTITYNQLEQAILDYAEGLRVIGV 130
S EWK VPDIWRSSAEKYGDK+AL+D YH PPST+TY QLE AILD+AEGLRVIGV
Sbjct: 334 ---VSDEWKTVPDIWRSSAEKYGDKIALIDPYHDPPSTMTYKQLEDAILDFAEGLRVIGV 504
Query: 131 RPDEKLALFADNSCRWLVADQGMM--ASGAINVVRGSRSSVEELLQ 174
P+EK+ALFADNSCRWLVADQ + ++ RSS+EELLQ
Sbjct: 505 SPNEKIALFADNSCRWLVADQV*V*WQPEQSMLLGVQRSSIEELLQ 642
Score = 38.5 bits (88), Expect = 0.009
Identities = 19/31 (61%), Positives = 23/31 (73%)
Frame = +3
Query: 152 GMMASGAINVVRGSRSSVEELLQIYNHSESV 182
GMMA+GAINVVRGS+ IYNHSE++
Sbjct: 573 GMMATGAINVVRGSKVIN*RTAAIYNHSEAL 665
>TC78604 similar to PIR|E84456|E84456 probable acyl-CoA synthetase [imported]
- Arabidopsis thaliana, partial (91%)
Length = 2573
Score = 153 bits (386), Expect = 3e-37
Identities = 155/642 (24%), Positives = 274/642 (42%), Gaps = 28/642 (4%)
Frame = +2
Query: 110 TYNQLEQAILDYAEGLRVIGVRPDEKLALFADNSCRWLVADQGMMASGAINVVRGSRSSV 169
TY ++ + ++A GL +G D +A+F+D W +A QG V +
Sbjct: 488 TYGEVFARVSNFASGLLKLGHDIDSHVAIFSDTRAEWFIALQGCFRQNITVVTIYASLGE 667
Query: 170 EELLQIYNHSESVALAVDGPEMFNRIAKPFYSKTGMRFIILLWGEKSDLNLIAEE-NKEV 228
+ L+ N ++ L D ++ N++ T ++ II + + + +E +
Sbjct: 668 DALIHSLNETQVSTLICD-VKLLNKLDAIRSKLTSLQNIIYFEDDSKEEHTFSEGLSSNC 844
Query: 229 PIFSFMEVIDLGRESRMALSDSHEASQRYVYEAINSDDIATLIYTSGTTGNPKGVMLTHR 288
I SF EV LG+ES + S + + +A ++YTSG+TG PKGVM+TH
Sbjct: 845 KIASFDEVEKLGKESPVEPS------------LPSKNAVAVVMYTSGSTGLPKGVMITHG 988
Query: 289 NLLHQIKNLWDTVPAEVG--DRFLSMLPPWHAYERACEYFIFTCGIEQVYTTVRNLKD-- 344
N++ + +P +G D +L+ LP H +E A E + G+ Y + L D
Sbjct: 989 NIVATTAAVMTVIP-NLGSKDVYLAYLPLAHGFEMAAESVMLAAGVAIGYGSPMTLTDTS 1165
Query: 345 ---------DLGRYQPHYMISVPLVFETLYSGIQKQISTSPPVRKLVALTFIRVSLAYME 395
D+ +P + +VP + + + G+ K++ + K +L +
Sbjct: 1166 NKVKKGTKGDVTVLKPTLLTAVPAIIDRIRDGVVKKVEEKGGLAK---------NLFQIA 1318
Query: 396 YKRIYEGKCLTRNVKQPSIVNSMLDCLWARIIATILFPIHLLAIKFVYSKIHSAIGLS-K 454
YKR + ++ S L L+ ++ KI + +G + +
Sbjct: 1319 YKR-----------RLAAVKGSWLGAWGVE---------KLVWDTIIFKKIRTVLGGNLR 1438
Query: 455 AGISGGGSLPLEVDKFFE-AIGVKVQNGYGLTETSPVIAARRPRCNVIGSVGHPVQHTEF 513
+ GG L + +F +G + GYGLTET A +G VG P+
Sbjct: 1439 FMLCGGAPLSGDSQQFINICVGAPIGQGYGLTETFAGAAFSEADDYSVGRVGPPLPCCYI 1618
Query: 514 KVVDSETGEVL---PPGSKGILKVRGPPVMNGYYKNPLATNQA--LDKDG--WLNTGDLG 566
K+V E G L P +G + V G V GY+KN T++ +D+ G W TGD+G
Sbjct: 1619 KLVSWEEGGYLSSDKPMPRGEVVVGGFSVTAGYFKNQDKTDEVFKVDEKGVRWFYTGDIG 1798
Query: 567 WIAPHHSTGRSRNSSGVIVVDGRAKDTIVLSTGENVEPAELEEAAMRSSIIQQIVVIGQD 626
P G + + R KD + L GE + ++E A + I+V
Sbjct: 1799 RFHP----------DGCLEIIDRKKDIVKLQHGEYISLGKVEAALASGDYVDSIMVHADP 1948
Query: 627 -KRRLGAIIVPNSEEVLKVARELSI----IDSISSNVVSEEKVLNLIYKELKTWMSESPF 681
A++V + + + K A+E I + + + +VL I K K +
Sbjct: 1949 FHSYCVALVVASHQSLEKWAQETGIEYKDFPDLCNKPEAVTEVLQSISKAAKAAKLQKTE 2128
Query: 682 QIGPILLVNEPFTIDNGLMTPTMKIRRDRVVAKYKEQIDDLY 723
I L+ +P+T ++GL+T +K++R+++ AK+ + + LY
Sbjct: 2129 VPAKIKLLADPWTPESGLVTAALKLKREQLKAKFNDDLQKLY 2254
>TC77271 similar to PIR|T07929|T07929 probable long-chain-fatty-acid--CoA
ligase (EC 6.2.1.3) isoform 2 - rape, partial (94%)
Length = 2486
Score = 97.8 bits (242), Expect(2) = 1e-36
Identities = 95/381 (24%), Positives = 166/381 (42%), Gaps = 9/381 (2%)
Frame = +1
Query: 353 YMISVPLVFETLYSGIQKQISTSPPVRKLVALTFIRVSLAYMEYKRIYEGKCLTRNVKQP 412
+ + VP V + +YSG+ ++IS+ ++K T + +Y + + +G+
Sbjct: 1114 FSVXVPRVLDRVYSGLTQKISSGGFLKK----TLFNFAYSY-KLNNMKKGQ-------NH 1257
Query: 413 SIVNSMLDCLWARIIATILFPIHLLAIKFVYSKIHSAIGLS-KAGISGGGSLPLEVDKFF 471
++ + +LD K V+ K+ +G S + +SG L L V+ +
Sbjct: 1258 AVASPLLD-------------------KIVFDKVKQGLGGSVRLILSGAAPLSLHVESYL 1380
Query: 472 EAIG-VKVQNGYGLTETSPVIAARRPR-CNVIGSVGHPVQHTE--FKVVDSETGEVLPPG 527
+ V GYGLTET P +++G+VG PV + + + V + L
Sbjct: 1381 RVVTCAHVLQGYGLTETCAGTFVSLPNELDMLGTVGPPVPNVDACLESVPEMGYDALAST 1560
Query: 528 SKGILKVRGPPVMNGYYKNPLATNQALDKDGWLNTGDLGWIAPHHSTGRSRNSSGVIVVD 587
+G + V+G + +GYYK T + L DGW +TGD+G P+ G + +
Sbjct: 1561 PRGEICVKGDTLFSGYYKREDLTKEVL-VDGWFHTGDIGEWQPN----------GSMKII 1707
Query: 588 GRAKDTIVLSTGENVEPAELEEAAMRSSIIQQIVVIGQDKRRLGAIIVPNSEEVLKVARE 647
R K+ LS GE V LE + ++ I V G +V S+ L+ E
Sbjct: 1708 DRKKNIFKLSQGEYVAVENLENIYSQVPSVESIWVYGNSFEAFLVAVVNPSKPALEHWAE 1887
Query: 648 LSIIDSISSNVVSEEKVLNLIYKELKTWMSESPFQ----IGPILLVNEPFTIDNGLMTPT 703
+ I +++ + + I EL E + I + L PF ++ L+TPT
Sbjct: 1888 ENGISVDFNSLCGDSRAKGYILDELSKIGKEKKLKGFEFIKAVHLDPVPFDMERDLITPT 2067
Query: 704 MKIRRDRVVAKYKEQIDDLYK 724
K +R +++ Y+ IDD+YK
Sbjct: 2068 YKKKRPQLLKYYQNVIDDMYK 2130
Score = 74.3 bits (181), Expect(2) = 1e-36
Identities = 63/249 (25%), Positives = 113/249 (45%), Gaps = 10/249 (4%)
Frame = +2
Query: 83 DIWRSSAEKYGDKVAL-----VDQYHHPPSTITYNQLEQAILDYAEGLRVIGVRPDEKLA 137
D++R S EKY + L VD H TY ++ ++ +R G K
Sbjct: 305 DVFRLSVEKYPNNPMLGSREIVDGKHGKYKWQTYKEVYDMVIKVGNSIRSCGYGEGVKCG 484
Query: 138 LFADNSCRWLVADQGMMASGAINVVRGSRSSVEELLQIYNHSESVALAVDGPEMFNRIAK 197
++ NS W+++ + A G V + I +H+E V++A + + K
Sbjct: 485 IYGANSAEWIMSMEACNAHGLHCVPLYDTLGSGAIEFIISHAE-VSIAFAEEKKIPELLK 661
Query: 198 PFYSKTGMRFIILLWGEKSDLNLIAEENKEVPIFSFMEVIDLGRESRMALSDSHEASQRY 257
F + T I+ +G+ + E + I+S+ E + LG+ S +
Sbjct: 662 TFPNATKYLKTIVSFGKVTPEQKQEVEKFGLAIYSWTEFLQLGK------------SHSF 805
Query: 258 VYEAINSDDIATLIYTSGTTGNPKGVMLTHRN---LLHQIKNLWDTVPAEVGDR--FLSM 312
DI T++YTSGTTG+PKGV++++ + LL +K L ++V ++ ++ +LS
Sbjct: 806 DLPVKKRSDICTIMYTSGTTGDPKGVLISNESIITLLAGVKRLLESVNEKLTEKDVYLSY 985
Query: 313 LPPWHAYER 321
LP H ++R
Sbjct: 986 LPLAHIFDR 1012
>TC87211 similar to GP|13516481|dbj|BAB40450. AMP-binding protein
{Arabidopsis thaliana}, partial (52%)
Length = 1338
Score = 129 bits (324), Expect = 4e-30
Identities = 102/393 (25%), Positives = 177/393 (44%), Gaps = 11/393 (2%)
Frame = +3
Query: 342 LKDDLGRYQPHYMISVPLVFETLYSGIQKQISTSPPVRKLVALTFIRVSLAYMEYKRIYE 401
L DDL +P SVP ++ +Y+GI + TS +++ + + Y
Sbjct: 30 LMDDLAALRPTVFCSVPRLYNRIYAGIINAVKTSGGLKERL-------------FNAAYN 170
Query: 402 GKCLTRNVKQPSIVNSMLDCLWARIIATILFPIHLLAIKFVYSKIHSAI-GLSKAGISGG 460
K +Q + +W R+ V++KI + G + +SG
Sbjct: 171 AK------RQALLHGKNPSPMWDRL---------------VFNKIKEKLGGRVRLMVSGA 287
Query: 461 GSLPLEVDKFFE-AIGVKVQNGYGLTETSPVIAARRPRCNVIGSVGHPVQHTEFKVVDSE 519
L +V +F + G +V GYG+TET+ VI+ + G VG P E K+VD
Sbjct: 288 SPLSPDVMEFLKICFGGRVTEGYGMTETTCVISCIDNGDRLGGHVGSPNPACEIKLVDVP 467
Query: 520 TGEVL---PPGSKGILKVRGPPVMNGYYKNPLATNQALDKDGWLNTGDLG-WIAPHHSTG 575
P +G + VRGP + GYYK+ T + +D++GWL+TGD+G WIA
Sbjct: 468 EMNYTSDDQPNPRGEICVRGPIIFQGYYKDEAQTREVIDEEGWLHTGDIGTWIA------ 629
Query: 576 RSRNSSGVIVVDGRAKDTIVLSTGENVEPAELEEAAMRSSIIQQIVVIGQDKRRLGAIIV 635
G + + R K+ L+ GE + P ++E ++ + + Q + G +V
Sbjct: 630 -----GGRLKIIDRKKNIFKLAQGEYIAPEKIENVYVKCNFVAQCFIYGDSFNSSLVSVV 794
Query: 636 PNSEEVLKV-ARELSIIDSISSNVVSEEKVLNLIYKELKTWMSESPFQ----IGPILLVN 690
+V+K A +I+ + + + ++ + + E+ E+ + + LV
Sbjct: 795 SVDPDVMKAWAASQNIVYNDLTQLCNDPRAKAAVLAEMDAVGREAQLRGFEFAKAVFLVA 974
Query: 691 EPFTIDNGLMTPTMKIRRDRVVAKYKEQIDDLY 723
EPF ++NGL+TPTMKI+R + + + I D+Y
Sbjct: 975 EPFAMENGLLTPTMKIKRPQAKEYFGKAISDMY 1073
>TC89975 similar to PIR|G96530|G96530 probable acyl CoA synthetase
[imported] - Arabidopsis thaliana, partial (46%)
Length = 1251
Score = 104 bits (260), Expect = 1e-22
Identities = 104/396 (26%), Positives = 178/396 (44%), Gaps = 10/396 (2%)
Frame = +1
Query: 339 VRNLKDDLGRYQPHYMISVPLVFETLYSGIQKQISTSPPVRKLVALTFIRVSLAYMEYKR 398
VR L +D+ +P VP VF+ + +GI+ ++S++ ++ A +Y
Sbjct: 1 VRFLLEDVQTLKPTICCGVPRVFDRISAGIKSKVSSAGSLQS-----------ALFQYAY 147
Query: 399 IYEGKCLTRNVKQPSIVNSMLDCLWARIIATILFPIHLLAIKFVYSKIHSAIG-LSKAGI 457
Y+ + L + + Q A F + V+ K A+G + +
Sbjct: 148 NYKLRYLEKGLPQHK--------------AAPFFD------RLVFDKTKQALGGRVRILL 267
Query: 458 SGGGSLPLEVDKFFEA-IGVKVQNGYGLTET-SPVIAARRPRCNVIGSVGHPVQHTEFKV 515
S LP+ V++ IG + GYGLTE+ + A +++G+VG P+ E ++
Sbjct: 268 SRAAPLPIHVEECLRVTIGSTLSQGYGLTESCAGCFTAIGDVFSMMGTVGIPMTTIEARL 447
Query: 516 VD-SETG-EVLPPGSKGILKVRGPPVMNGYYKNPLATNQALDKDGWLNTGDLGWIAPHHS 573
E G + L +G + +RG + +GY+K T + + DGW +TGD+G P+
Sbjct: 448 ESVPEMGYDALSSEPRGEICLRGNSLFSGYHKRQDLTQEVI-VDGWFHTGDIGEWQPN-- 618
Query: 574 TGRSRNSSGVIVVDGRAKDTIVLSTGENVEPAELEEAAMRSSIIQQIVVIGQD-KRRLGA 632
G + + R K+ LS GE + +E ++ +I I V G + L A
Sbjct: 619 --------GAMKIIDRKKNIFKLSQGEYIAVENIESKYLQCPLISSIWVYGNSFESFLVA 774
Query: 633 IIVPNSEEVLKVARELSIIDSISSNVVSEEKVLNLIYKELKTWMSESPF----QIGPILL 688
++VP + + A E + S V K I ELK+ + Q+ I L
Sbjct: 775 VVVPERKALEDWAVEHNFTGDFQS-VCENLKARQYILDELKSTGQKLQLRGFEQLKAIYL 951
Query: 689 VNEPFTIDNGLMTPTMKIRRDRVVAKYKEQIDDLYK 724
PF I+ L+TPT K++R +++ YK+QID+LYK
Sbjct: 952 EPSPFDIERDLVTPTFKLKRPQLLKYYKDQIDELYK 1059
>TC86993 similar to GP|13516481|dbj|BAB40450. AMP-binding protein
{Arabidopsis thaliana}, partial (65%)
Length = 1523
Score = 95.9 bits (237), Expect = 5e-20
Identities = 93/388 (23%), Positives = 155/388 (38%), Gaps = 4/388 (1%)
Frame = +2
Query: 109 ITYNQLEQAILDYAEGLRVIGVRPDEKLALFADNSCRWLVADQGMMASGAINVVRGSRSS 168
+TY + A GL G+ + L+ N WL+ D A I+V
Sbjct: 416 MTYGEAGTARSAIGSGLIHYGIPKGAGIGLYFINRPEWLIVDHACSAYSYISVPLYDTLG 595
Query: 169 VEELLQIYNHSESVALAVDGPEMFNRIAKPFYSKTGMRFIILLWGEKSDLNLIAEENKEV 228
+ + I NH+ V + + N + +R I+++ G + + + V
Sbjct: 596 PDAVKYIVNHA-LVQVIFCVSQTLNSLLSYLSEIPTVRLIVVVGGIDDQIPSLPSSDG-V 769
Query: 229 PIFSFMEVIDLGRESRMALSDSHEASQRYVYEAINSDDIATLIYTSGTTGNPKGVMLTHR 288
I S+ ++ GR + +D+AT+ YTSGTTG PKG +LTH
Sbjct: 770 QIISYTKLFSQGRSNLQPFCPP------------KPEDVATICYTSGTTGTPKGAVLTHE 913
Query: 289 NLLHQIKNLWDTVPAEVGDRFLSMLPPWHAYERACEYFIFTCGIEQVYTTVRNLK--DDL 346
N + + D ++S LP H YERA + G+ + NLK DDL
Sbjct: 914 NFIANVAGATIDEKFNPSDVYISYLPLAHIYERANQVMTVYFGMAVGFYQGDNLKLMDDL 1093
Query: 347 GRYQPHYMISVPLVFETLYSGIQKQISTSPPVRKLVALTFIRVSLAYMEYKRIYEGKCLT 406
+P SVP ++ +Y+GI + TS +++ + + Y K
Sbjct: 1094AALRPTVFCSVPRLYNRIYAGIINAVKTSGGLKERL-------------FNAAYNAK--- 1225
Query: 407 RNVKQPSIVNSMLDCLWARIIATILFPIHLLAIKFVYSKIHSAI-GLSKAGISGGGSLPL 465
+Q + +W R+ V++KI + G + +SG L
Sbjct: 1226---RQALLHGKNPSPMWDRL---------------VFNKIKEKLGGRVRLMVSGASPLSP 1351
Query: 466 EVDKFFE-AIGVKVQNGYGLTETSPVIA 492
+V +F + G +V GYG+TET+ VI+
Sbjct: 1352DVMEFLKICFGGRVTEGYGMTETTCVIS 1435
>TC77196 similar to GP|17063848|gb|AAL35216.1 4-coumarate:CoA ligase {Amorpha
fruticosa}, partial (93%)
Length = 1924
Score = 89.7 bits (221), Expect = 3e-18
Identities = 76/264 (28%), Positives = 114/264 (42%), Gaps = 7/264 (2%)
Frame = +3
Query: 458 SGGGSLPLEVDKFFEAI--GVKVQNGYGLTETSPVIA-----ARRPRCNVIGSVGHPVQH 510
SGG L E++ A K+ GYG+TE PV+ A+ P G+ G V++
Sbjct: 1035 SGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDVKSGACGTVVRN 1214
Query: 511 TEFKVVDSETGEVLPPGSKGILKVRGPPVMNGYYKNPLATNQALDKDGWLNTGDLGWIAP 570
E K+VD + LP G + +RG +M GY NP AT + +DK+GWL+TGD+G+I
Sbjct: 1215 AEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIGFI-- 1388
Query: 571 HHSTGRSRNSSGVIVVDGRAKDTIVLSTGENVEPAELEEAAMRSSIIQQIVVIGQDKRRL 630
+ + +VD R K+ ++ G V PAELE + I + V+
Sbjct: 1389 -------DDDDELFIVD-RLKE-LIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAA 1541
Query: 631 GAIIVPNSEEVLKVARELSIIDSISSNVVSEEKVLNLIYKELKTWMSESPFQIGPILLVN 690
G + V V R ID+ ++ + YK +N
Sbjct: 1542 GEV------PVAFVVRSNGSIDTTEDDIKKFVSKQVVFYKR-----------------IN 1652
Query: 691 EPFTIDNGLMTPTMKIRRDRVVAK 714
F ID +P+ KI R + AK
Sbjct: 1653 RVFFIDAIPKSPSGKILRKDLRAK 1724
Score = 52.0 bits (123), Expect = 8e-07
Identities = 62/237 (26%), Positives = 94/237 (39%), Gaps = 8/237 (3%)
Frame = +1
Query: 91 KYGDKVALVDQYHHPPSTI-TYNQLEQAILDYAEGLRVIGVRPDEKLALFADNSCRWLVA 149
KYG + L++ P + I TY ++ A GL +G++ + + + N ++ A
Sbjct: 220 KYGSRPCLINA---PTAEIYTYYDVQLTAQKVASGLNKLGIQQGDVIMVLLPNCPEFVFA 390
Query: 150 DQGMMASGAINVVRGSRSSVEELLQIYNHSESVALAVDGPEMFNRIAKPFYSKTGMRFII 209
G GAI + E+ + S + L +Y K
Sbjct: 391 FLGASFRGAIMTAANPFFTSAEIAKQAKASNTKLLVTQAC---------YYDKV------ 525
Query: 210 LLWGEKSDLNLIAEENKEVPIFSFMEVIDLGRESRMALSDSHEASQRYVYEA---INSDD 266
DL E K V + S E ++ M S+ +A Q + E I DD
Sbjct: 526 ------KDL----ENVKLVFVDSSPE-----EDNHMHFSELIQADQNEMEEVKVNIKPDD 660
Query: 267 IATLIYTSGTTGNPKGVMLTHRNLLHQIKNLWD----TVPAEVGDRFLSMLPPWHAY 319
+ L Y+SGTTG PKGVMLTH+ L+ I D + D L +LP +H Y
Sbjct: 661 VVALPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCVLPMFHIY 831
>TC81027 similar to PIR|H85064|H85064 4-coumarate--CoA ligase-like protein
[imported] - Arabidopsis thaliana, partial (28%)
Length = 1283
Score = 88.6 bits (218), Expect = 8e-18
Identities = 60/184 (32%), Positives = 89/184 (47%), Gaps = 8/184 (4%)
Frame = +1
Query: 460 GGSLPLEVDKFFEAIG----VKVQNGYGLTETSPVIAARRPR----CNVIGSVGHPVQHT 511
GG+ PL D E V V GYG+TE +++ P+ +V GS G +
Sbjct: 343 GGAAPLGKDLMQECTKILPHVHVIQGYGMTEACGLVSIENPKEGSLISVSGSTGTLIPSV 522
Query: 512 EFKVVDSETGEVLPPGSKGILKVRGPPVMNGYYKNPLATNQALDKDGWLNTGDLGWIAPH 571
E ++++ T + LPP G + +RGP +M GY+ NP AT A++ GW+ TGDLG+
Sbjct: 523 ESRIINLATLKPLPPNQLGEIWLRGPTIMQGYFNNPEATKLAINDQGWMITGDLGYF--- 693
Query: 572 HSTGRSRNSSGVIVVDGRAKDTIVLSTGENVEPAELEEAAMRSSIIQQIVVIGQDKRRLG 631
+ G + V R K+ I S G V PAELE+ + I VI ++G
Sbjct: 694 -------DEKGQLFVVDRIKELIKCS-GYQVAPAELEDLLVSHPEISDAGVIPSPDAKVG 849
Query: 632 AIIV 635
+ V
Sbjct: 850 EVPV 861
>TC77753 similar to GP|19773582|gb|AAL98709.1 4-coumarate:coenzyme A ligase
{Glycine max}, partial (94%)
Length = 2034
Score = 85.5 bits (210), Expect = 7e-17
Identities = 64/215 (29%), Positives = 104/215 (47%), Gaps = 6/215 (2%)
Frame = +3
Query: 427 IATILFPIHLLAIKFVYSKIHSAIGLSKAGISGGGSLPLEVDKFFE--AIGVKVQNGYGL 484
+A+ + PI L +K S + + + I+G + +E+++ + + GYG+
Sbjct: 903 VASFVPPIVLALVKSGESMRYDLSSI-RVMITGAAPMGMELEQAVKDRLPRTVLGQGYGM 1079
Query: 485 TETSPVIA----ARRPRCNVIGSVGHPVQHTEFKVVDSETGEVLPPGSKGILKVRGPPVM 540
TE P+ A+ P G+ G V++ E K+VD+ETG LP G + +RG VM
Sbjct: 1080 TEAGPLSISLAFAKEPFKTKPGACGTVVRNAEMKIVDTETGASLPRNKAGEICIRGTKVM 1259
Query: 541 NGYYKNPLATNQALDKDGWLNTGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDTIVLSTGE 600
GY +P AT + +DK+GWL+TGD+G I + + +VD R K+ ++ G
Sbjct: 1260 KGYLNDPEATKRTIDKEGWLHTGDIGLI---------DDDDELFIVD-RLKE-LIKYKGY 1406
Query: 601 NVEPAELEEAAMRSSIIQQIVVIGQDKRRLGAIIV 635
V PAELE + I V+ G + V
Sbjct: 1407 QVAPAELEALLIAHPNISDAAVVPLKDEAAGEVPV 1511
Score = 55.1 bits (131), Expect = 1e-07
Identities = 57/229 (24%), Positives = 95/229 (40%), Gaps = 4/229 (1%)
Frame = +3
Query: 108 TITYNQLEQAILDYAEGLRVIGVRPDEKLALFADNSCRWLVADQGMMASGAINVVRGSRS 167
T+TY+ + + A GL +G++ + + + NS ++ + G GA+
Sbjct: 219 TLTYSDVHLTVRKIAAGLNTLGIQQGDVIMIVLRNSPQFALTFLGASFRGAVITTANPFY 398
Query: 168 SVEELLQIYNHSESVALAVDGPEMFNRIAKPFYSKTGMRFIILLWGEKSDLNLIAEENKE 227
+ EL + ++S L + N+I F ++ + + + D N
Sbjct: 399 TSSELAKQATATKS-KLIITQSVYLNKI-NDFAKLIDIKIVCIDSSPEEDEN-------- 548
Query: 228 VPIFSFMEVIDLGRESRMALSDSHEASQRYVYEAINSDDIATLIYTSGTTGNPKGVMLTH 287
V+D S + +D +E + IN +D+ L ++SGT+G PKGVMLTH
Sbjct: 549 --------VVDF---SVLTNADENELPE----VKINPNDVVALPFSSGTSGLPKGVMLTH 683
Query: 288 RNLLHQIKNLWDTVP----AEVGDRFLSMLPPWHAYERACEYFIFTCGI 332
NL+ I L D D L +LP +H Y I CGI
Sbjct: 684 ENLVTTISQLVDGENPHQYTNYEDVLLCVLPMFHIYALNS---ILLCGI 821
>BI311229 similar to GP|20502991|gb Putative AMP-binding protein {Oryza
sativa (japonica cultivar-group)}, partial (29%)
Length = 693
Score = 84.7 bits (208), Expect = 1e-16
Identities = 72/222 (32%), Positives = 105/222 (46%), Gaps = 8/222 (3%)
Frame = +3
Query: 456 GISGGGSLPLEVDKFFEAIGVKVQNGYGLTE-TSPVIAARRPR-CNVIGSVGHPVQHTEF 513
G G G+ + KF A + GYGLTE T+ VI P + G+ G V E
Sbjct: 3 GSVGKGNFVAFMAKFPHA---SIIQGYGLTESTAGVIRIVGPEEASRGGTTGKLVSGMEA 173
Query: 514 KVVDSETGEVLPPGSKGILKVRGPPVMNGYYKNPLATNQALDKDGWLNTGDLGWIAPHHS 573
K+V+ TGE + PG +G L VRGPP+M GY +P+AT+ L DGWL TGD+ +
Sbjct: 174 KIVNPNTGEAMSPGEQGELWVRGPPIMKGYVGDPVATSVTL-VDGWLRTGDICYF----- 335
Query: 574 TGRSRNSSGVIVVDGRAKDTIVLSTGENVEPAELEEAAMRSSIIQQIVVIGQDKRRLGAI 633
++ G + V R K+ ++ G V PAELE+ I+ VI G I
Sbjct: 336 -----DNEGFVYVVDRLKE-LIKYKGYQVAPAELEQLLQSHPEIKDAAVIPYPDEDAGQI 497
Query: 634 IV------PNSEEVLKVARELSIIDSISSNVVSEEKVLNLIY 669
+ P+S E II+ ++ V +KV +++
Sbjct: 498 PLAFVIRQPHSS-----MGEAEIINFVAKQVAPYKKVRRVVF 608
>TC88054 homologue to PIR|PQ0772|PQ0772 4-coumarate--CoA ligase (EC
6.2.1.12) (clone GM4CL1B) - soybean (fragment), partial
(62%)
Length = 1055
Score = 82.8 bits (203), Expect = 4e-16
Identities = 55/160 (34%), Positives = 80/160 (49%), Gaps = 5/160 (3%)
Frame = +1
Query: 481 GYGLTETSPVIA-----ARRPRCNVIGSVGHPVQHTEFKVVDSETGEVLPPGSKGILKVR 535
GYG+TE PV++ A+ P GS G V++ E KV+D ETG L G + +R
Sbjct: 178 GYGMTEAGPVLSMSLGFAKNPFPTSSGSCGTVVRNAELKVLDPETGRSLGYNQPGEICIR 357
Query: 536 GPPVMNGYYKNPLATNQALDKDGWLNTGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDTIV 595
G +M GY + AT +D++GWL+TGD+G+I ++ + +VD R K+ I
Sbjct: 358 GQQIMKGYLNDENATKTTIDEEGWLHTGDVGYI---------DDNDEIFIVD-RVKELIK 507
Query: 596 LSTGENVEPAELEEAAMRSSIIQQIVVIGQDKRRLGAIIV 635
G V PAELE + I V+ Q G + V
Sbjct: 508 FK-GFQVPPAELEGLLVSHPSIADAAVVPQKDVAAGEVPV 624
>AW329094 similar to GP|20502991|gb Putative AMP-binding protein {Oryza
sativa (japonica cultivar-group)}, partial (34%)
Length = 595
Score = 81.3 bits (199), Expect = 1e-15
Identities = 61/188 (32%), Positives = 87/188 (45%), Gaps = 5/188 (2%)
Frame = +2
Query: 476 VKVQNGYGLTETSPVIAARRP--RCNVIGSVGHPVQHTEFKVVDSETGEVLPPGSKGILK 533
V + GYGLTE+S + A+ G+ G TE +VD+ET + LP G L
Sbjct: 59 VSIFQGYGLTESSGIGASTESLEESRKYGTAGLVSASTEAMIVDTETAQPLPVNRTGELW 238
Query: 534 VRGPPVMNGYYKNPLATNQALDKDGWLNTGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDT 593
+RGP M GY+ N AT + +GWL TGD+ +I +S G + V R K+
Sbjct: 239 LRGPTTMKGYFSNEEATRSTITPEGWLKTGDVCYI----------DSDGFLFVVDRLKE- 385
Query: 594 IVLSTGENVEPAELEEAAMRSSIIQQIVVIGQDKRRLGAIIVPNSEEVLKVARELS---I 650
++ G V PAELE + I VI + G VP + V V LS +
Sbjct: 386 LIKYKGYQVPPAELEALLLTHPAILDAAVIPYPDKEAGQ--VPMAYVVRNVGSNLSGSQV 559
Query: 651 IDSISSNV 658
+D ++ V
Sbjct: 560 MDFVAEQV 583
>AW776080 similar to GP|20161027|db putative 4-coumarate-CoA ligase {Oryza
sativa (japonica cultivar-group)}, partial (28%)
Length = 625
Score = 79.0 bits (193), Expect = 6e-15
Identities = 55/166 (33%), Positives = 80/166 (48%), Gaps = 3/166 (1%)
Frame = +2
Query: 502 GSVGHPVQHTEFKVVDSETGEVLPPGSKGILKVRGPPVMNGYYKNPLATNQALDKDGWLN 561
GSVG ++ E K+VD TGE L PG KG L +RGP +M GY + AT + LD +GWL
Sbjct: 65 GSVGRLAENMEAKIVDPVTGEALSPGQKGELWLRGPTIMKGYVGDDKATVETLDSEGWLK 244
Query: 562 TGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDTIVLSTGENVEPAELEEAAMRSSIIQQIV 621
TGDL + +S G + + R K+ I + V PAELE + I
Sbjct: 245 TGDLCYF----------DSDGYLFIVDRLKELIKYKAYQ-VPPAELEHILHTNPEIADAA 391
Query: 622 VIGQDKRRLGAIIVPNSEEVLKVARELS---IIDSISSNVVSEEKV 664
V+ G I P + V K ++ ++D ++ V +K+
Sbjct: 392 VVPYPDEDAGQI--PMAFVVRKPGSNITAAQVMDYVAKQVTPYKKI 523
>BI269977 similar to GP|12039389|gb putative 4-coumarate CoA ligase {Oryza
sativa}, partial (28%)
Length = 520
Score = 78.6 bits (192), Expect = 8e-15
Identities = 60/182 (32%), Positives = 86/182 (46%), Gaps = 5/182 (2%)
Frame = +1
Query: 482 YGLTETSPVIAA--RRPRCNVIGSVGHPVQHTEFKVVDSETGEVLPPGSKGILKVRGPPV 539
YG+TE+ V + + S+G + E KVVD ++G LPPG G L +RGP +
Sbjct: 1 YGMTESGAVGTRGFNTDKFHNYSSLGLLAPNIEAKVVDWKSGTFLPPGLSGELWLRGPSI 180
Query: 540 MNGYYKNPLATNQALDKDGWLNTGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDTIVLSTG 599
M GY N AT +DKDGW++TGD+ + + G + + GR KD I+ G
Sbjct: 181 MKGYLNNEEATMSTIDKDGWIHTGDIVYF----------DQDGYLYMSGRLKD-IIKYKG 327
Query: 600 ENVEPAELEEAAMRSSIIQQIVVIGQDKRRLGAIIVPNSEEVLKVARELS---IIDSISS 656
+ PA+LE + I V G I P + V KV LS +ID ++
Sbjct: 328 FQIAPADLEALLISHPEIVDAAVTAGKVDVAGEI--PVAFVVKKVGSVLSSQHVIDYVAX 501
Query: 657 NV 658
V
Sbjct: 502 QV 507
>TC91703 similar to GP|4038975|gb|AAC97600.1| 4-coumarate:CoA ligase
isoenzyme 2 {Glycine max}, partial (48%)
Length = 828
Score = 75.5 bits (184), Expect = 7e-14
Identities = 45/121 (37%), Positives = 66/121 (54%), Gaps = 10/121 (8%)
Frame = +1
Query: 458 SGGGSLPLEVD-----KFFEAIGVKVQNGYGLTETSPVIA-----ARRPRCNVIGSVGHP 507
SGG L E++ KF AI + GY +TE PV+ A+ P + G
Sbjct: 421 SGGAPLGKELEDTVRTKFPNAI---LGQGYRMTEAGPVLTMSLAFAKEPLNVKASACGTV 591
Query: 508 VQHTEFKVVDSETGEVLPPGSKGILKVRGPPVMNGYYKNPLATNQALDKDGWLNTGDLGW 567
V++ E K+VD +TG+ LP G + +RG +M GY + AT + +DK+GWL TGD+G+
Sbjct: 592 VRNAEMKIVDPDTGKSLPRNQSGEICIRGDQIMKGYLNDLEATERTIDKEGWLYTGDIGY 771
Query: 568 I 568
I
Sbjct: 772 I 774
Score = 49.3 bits (116), Expect = 5e-06
Identities = 32/92 (34%), Positives = 44/92 (47%), Gaps = 4/92 (4%)
Frame = +1
Query: 262 INSDDIATLIYTSGTTGNPKGVMLTHRNLLHQIKNLWD----TVPAEVGDRFLSMLPPWH 317
I DD+ L Y+SGTTG PKGVMLTH+ L+ I D + D L +LP +H
Sbjct: 34 IQPDDVVALPYSSGTTGLPKGVMLTHKGLVSSIAQQVDGENPNLYYRSEDVILCVLPLFH 213
Query: 318 AYERACEYFIFTCGIEQVYTTVRNLKDDLGRY 349
Y + CG+ T + K D+ +
Sbjct: 214 IYSLNS---VLLCGLRAKATILLMPKFDINSF 300
>AL389862 similar to GP|23315116|gb Sequence 281 from patent US 6410718,
partial (90%)
Length = 535
Score = 70.9 bits (172), Expect = 2e-12
Identities = 51/171 (29%), Positives = 80/171 (45%)
Frame = +1
Query: 502 GSVGHPVQHTEFKVVDSETGEVLPPGSKGILKVRGPPVMNGYYKNPLATNQALDKDGWLN 561
G+ G V E ++V +T + LPP G + +RGP +M GY+ NP AT Q ++ GW
Sbjct: 10 GAAGSSV---ESQIVSLQTSKSLPPNQLGEIWLRGPVMMQGYFNNPEATKQTINDQGWTL 180
Query: 562 TGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDTIVLSTGENVEPAELEEAAMRSSIIQQIV 621
TGDLG+ + G + V R K+ ++ G V PAELE+ + I
Sbjct: 181 TGDLGYF----------DEKGQLFVVDRIKE-LIKCNGYQVAPAELEDLLISHPEISDAG 327
Query: 622 VIGQDKRRLGAIIVPNSEEVLKVARELSIIDSISSNVVSEEKVLNLIYKEL 672
VI + G + V V R L +++++EE + + KE+
Sbjct: 328 VIPSPDAKAGEV------PVAFVVRSL-------NSLITEEDIKKFVAKEV 441
>BG588502 SP|P31552|CAIC Probable crotonobetaine/carnitine-CoA ligase (EC
6.3.2.-). {Escherichia coli}, partial (50%)
Length = 807
Score = 58.2 bits (139), Expect(2) = 2e-11
Identities = 53/171 (30%), Positives = 75/171 (42%), Gaps = 7/171 (4%)
Frame = -1
Query: 488 SPVIAARRPRCNVIGSVGHPVQHTEFKVVDSETGEVLPPGSKGILKVRGPP---VMNGYY 544
S ++ A + R IG VG + + + LP G G + ++G P + Y+
Sbjct: 474 SAIVLANKRRWPSIGRVGFCYEAE----IRDDHNRPLPAGEIGEICIKGIPGKTIFKEYF 307
Query: 545 KNPLATNQALDKDGWLNTGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDTIVLSTGENVEP 604
NP AT + L+ DGWL+TGD TG VD R ++ GENV
Sbjct: 306 LNPQATAKVLEADGWLHTGD---------TGYRDEEDFFYFVDRRC--NMIKRGGENVSC 160
Query: 605 AELEEAAMRSSIIQQIVVIG-QDKRR---LGAIIVPNSEEVLKVARELSII 651
ELE IQ IVV+G +D R + A +V N E L E S++
Sbjct: 159 VELENIIAAHPKIQDIVVVGIKDSIRDEAIKAFVVLNEGETLS-EEEFSVL 10
Score = 28.9 bits (63), Expect(2) = 2e-11
Identities = 14/31 (45%), Positives = 17/31 (54%)
Frame = -3
Query: 466 EVDKFFEAIGVKVQNGYGLTETSPVIAARRP 496
E D F E GV++ YG+TET I RP
Sbjct: 553 EKDAFCERFGVRLLTSYGMTETIVGIIGDRP 461
>TC91955 weakly similar to PIR|T27421|T27421 hypothetical protein Y76A2B.3 -
Caenorhabditis elegans, partial (7%)
Length = 894
Score = 62.0 bits (149), Expect = 8e-10
Identities = 60/237 (25%), Positives = 94/237 (39%), Gaps = 32/237 (13%)
Frame = +2
Query: 110 TYNQLEQAILDYAEGLRVIGVRPDEK----------------LALFADNSCRWLVADQGM 153
TY Q+ + I ++ GL + ++ + ++A+N W + DQ
Sbjct: 197 TYKQIAERIDNFGSGLLYLNENTNKNPKVKKNPKNFKDKQWTVGIYANNRPEWFITDQAN 376
Query: 154 MASGAINVVRGSRSSVEELLQIYNHSESVALAVDGPEMFNRIAKPFYSKTGMRFIILLWG 213
A I V E + + NH+E + + V G + + SK +I+
Sbjct: 377 CAYNLITVALYDTXGPETVEFVINHAE-IPIVVTGASRIPGLIQ-LASKVPNLKVIISMD 550
Query: 214 EKSDLNLIA-------------EENKEVPIFSFMEVIDLGRESRMALSDSHEASQRYVYE 260
E D + + E+K + + SF EV LG + +
Sbjct: 551 ELEDDSPVPFGSTTTGKVLKAWAEDKGIVLLSFSEVEKLGNQYPRK------------HN 694
Query: 261 AINSDDIATLIYTSGTTGNPKGVMLTHRNLLHQIKN---LWDTVPAEVGDRFLSMLP 314
+ D+A + YTSGTTG PKGVMLTHRN + I + LWD D +S LP
Sbjct: 695 PPSPKDLACICYTSGTTGVPKGVMLTHRNFVAAISSSCQLWD---GNQDDVLISYLP 856
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.136 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,757,198
Number of Sequences: 36976
Number of extensions: 303477
Number of successful extensions: 1920
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 1648
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1806
length of query: 724
length of database: 9,014,727
effective HSP length: 103
effective length of query: 621
effective length of database: 5,206,199
effective search space: 3233049579
effective search space used: 3233049579
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146585.9


