

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146585.12 + phase: 0 /pseudo
(942 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
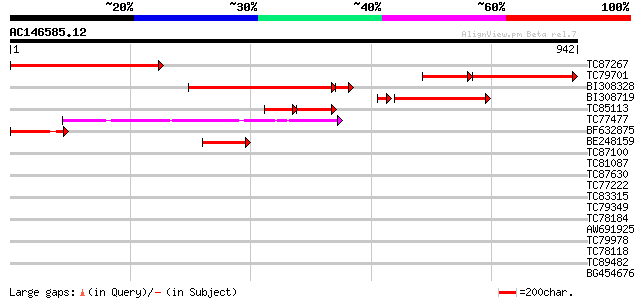
Score E
Sequences producing significant alignments: (bits) Value
TC87267 similar to PIR|T06242|T06242 aspartate kinase (EC 2.7.2.... 499 e-141
TC79701 homologue to PIR|T06242|T06242 aspartate kinase (EC 2.7.... 343 e-140
BI308328 similar to PIR|T06242|T06 aspartate kinase (EC 2.7.2.4)... 410 e-124
BI308719 similar to PIR|T06242|T06 aspartate kinase (EC 2.7.2.4)... 295 2e-83
TC85113 homologue to PIR|T06242|T06242 aspartate kinase (EC 2.7.... 119 7e-52
TC77477 homologue to GP|5305740|gb|AAD41796.1| precursor monofun... 179 4e-45
BF632875 similar to PIR|T06242|T062 aspartate kinase (EC 2.7.2.4... 177 2e-44
BE248159 homologue to PIR|T06246|T062 aspartate kinase (EC 2.7.2... 164 1e-40
TC87100 homologue to GP|9293891|dbj|BAB01794.1 contains similari... 36 0.079
TC81087 homologue to GP|530207|gb|AAA66338.1|| heat shock protei... 32 1.1
TC87630 similar to GP|11385453|gb|AAG34809.1 glutathione S-trans... 31 2.0
TC77222 weakly similar to GP|19911209|dbj|BAB86931. glucosyltran... 30 3.3
TC83315 weakly similar to GP|19911209|dbj|BAB86931. glucosyltran... 30 3.3
TC79349 similar to PIR|S65668|S65668 preprotein translocase secA... 30 4.3
TC78184 similar to GP|17979207|gb|AAL49842.1 unknown protein {Ar... 30 5.7
AW691925 30 5.7
TC79978 weakly similar to GP|20197257|gb|AAC28988.2 putative ABC... 29 7.4
TC78118 similar to PIR|T06528|T06528 lectin - garden pea, partia... 29 7.4
TC89482 similar to SP|Q9M3H5|AHM1_ARATH Potential cadmium/zinc-t... 29 9.7
BG454676 29 9.7
>TC87267 similar to PIR|T06242|T06242 aspartate kinase (EC 2.7.2.4) /
homoserine dehydrogenase (EC 1.1.1.3) precursor -
soybean, partial (24%)
Length = 817
Score = 499 bits (1285), Expect = e-141
Identities = 253/255 (99%), Positives = 253/255 (99%)
Frame = +3
Query: 1 MASSLSSSLSHFSRISVTSLQHDYNNKIPADSQCRHFLLSRRFHSLRKGITLPRRRESPS 60
MASSLSSSLSHFSRISVTSLQHDYNNKIPADSQCRHFLLSRRFHSLRKGITLPRRRESPS
Sbjct: 51 MASSLSSSLSHFSRISVTSLQHDYNNKIPADSQCRHFLLSRRFHSLRKGITLPRRRESPS 230
Query: 61 SGICASLTDVSVNVAVEEKELSKGDSWSVHKFGGTCMGSSQRIKNVGDIVLNDDSERKLV 120
SGICASLTDVSVNVAVEEKELSKGDSWSVHKFGGTCMGSSQRIKNVGDIVLNDDSERKLV
Sbjct: 231 SGICASLTDVSVNVAVEEKELSKGDSWSVHKFGGTCMGSSQRIKNVGDIVLNDDSERKLV 410
Query: 121 VVSAMSKVTDMMYDLINKAQSRDESYISSLDAVLEKHSATAHDILDGETLAIFLSKLHED 180
VVSAMSKVTDMMYDLINKAQSRDESYISSLDAVLEKHSATAHDILDGETLAIFLSKLHED
Sbjct: 411 VVSAMSKVTDMMYDLINKAQSRDESYISSLDAVLEKHSATAHDILDGETLAIFLSKLHED 590
Query: 181 ISNLKAMLRAIYIAGHVTESFTDFVVGHGELWSAQMLSLVIRKNGIDCKWMDTREVLIVN 240
ISNLKAMLRAIYIAGHVTESFTDFVVGHGELWSAQMLSLVIRKNGIDCKWMDTREVLIVN
Sbjct: 591 ISNLKAMLRAIYIAGHVTESFTDFVVGHGELWSAQMLSLVIRKNGIDCKWMDTREVLIVN 770
Query: 241 PTSSNQVDPDYLESE 255
PTS NQVDPDY ESE
Sbjct: 771 PTSXNQVDPDYXESE 815
>TC79701 homologue to PIR|T06242|T06242 aspartate kinase (EC 2.7.2.4) /
homoserine dehydrogenase (EC 1.1.1.3) precursor -
soybean, partial (28%)
Length = 1042
Score = 343 bits (880), Expect(2) = e-140
Identities = 174/174 (100%), Positives = 174/174 (100%)
Frame = +1
Query: 769 KDGRAFSEVVGEAKEAGYTEPDPRDDLSGTDVARKVIILARESGLKLELSNIPIESLVPE 828
KDGRAFSEVVGEAKEAGYTEPDPRDDLSGTDVARKVIILARESGLKLELSNIPIESLVPE
Sbjct: 253 KDGRAFSEVVGEAKEAGYTEPDPRDDLSGTDVARKVIILARESGLKLELSNIPIESLVPE 432
Query: 829 PLRACASAQEFMQQLPKFDQEFAKKQEDADNAGEVLRYVGVVDVTNKKGVVELRKYKKDH 888
PLRACASAQEFMQQLPKFDQEFAKKQEDADNAGEVLRYVGVVDVTNKKGVVELRKYKKDH
Sbjct: 433 PLRACASAQEFMQQLPKFDQEFAKKQEDADNAGEVLRYVGVVDVTNKKGVVELRKYKKDH 612
Query: 889 PFAQLSGSDNIIAFTTRRYKNQPLIVRGPGAGAQVTAGGIFSDILRLASYLGAP 942
PFAQLSGSDNIIAFTTRRYKNQPLIVRGPGAGAQVTAGGIFSDILRLASYLGAP
Sbjct: 613 PFAQLSGSDNIIAFTTRRYKNQPLIVRGPGAGAQVTAGGIFSDILRLASYLGAP 774
Score = 174 bits (441), Expect(2) = e-140
Identities = 84/84 (100%), Positives = 84/84 (100%)
Frame = +3
Query: 686 YEWLCKGIHVITPNKKANSGPLEQYLRLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLE 745
YEWLCKGIHVITPNKKANSGPLEQYLRLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLE
Sbjct: 3 YEWLCKGIHVITPNKKANSGPLEQYLRLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLE 182
Query: 746 TGDKILQIEGIFSGTLSYIFNNFK 769
TGDKILQIEGIFSGTLSYIFNNFK
Sbjct: 183 TGDKILQIEGIFSGTLSYIFNNFK 254
>BI308328 similar to PIR|T06242|T06 aspartate kinase (EC 2.7.2.4) /
homoserine dehydrogenase (EC 1.1.1.3) precursor -
soybean, partial (28%)
Length = 833
Score = 410 bits (1053), Expect(2) = e-124
Identities = 207/245 (84%), Positives = 226/245 (91%)
Frame = +2
Query: 298 IMGSLFRARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIP 357
+ G++FRARQVTIWTDVDGVYSADPRKVS+AVILKTLSYQEAWEMSYFGANVLHPRTI P
Sbjct: 8 LWGAIFRARQVTIWTDVDGVYSADPRKVSDAVILKTLSYQEAWEMSYFGANVLHPRTISP 187
Query: 358 VMRYGIPILIRNIFNLSAPGTKICHPVVSDYEDKSNLQNYVKGFATIDNLALVNVEGTGM 417
V+RYGIPI+IRNIFN SA GTKICHP + + EDK L++YVKGF TIDNLALVNVEGTGM
Sbjct: 188 VIRYGIPIIIRNIFNTSASGTKICHPSIIENEDKKILKDYVKGFTTIDNLALVNVEGTGM 367
Query: 418 AGVPGTASAIFAAVKDVGANVIMISQASSEHSVCFAVPEKEVKAVAEALQSRFRQALDNG 477
AGVPGTAS IF+AVK+VGANVIMISQASSEHSVCFAVPEKEVKAVAE L+S F AL G
Sbjct: 368 AGVPGTASTIFSAVKEVGANVIMISQASSEHSVCFAVPEKEVKAVAEVLESIFDSALFAG 547
Query: 478 RLSQVAVIPNCSILAAVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVIKR 537
R+SQVAVI NCSILAAVGQKMASTPGVSATLFNALA+ANIN+ AIAQGCSEYN+TVV+KR
Sbjct: 548 RISQVAVISNCSILAAVGQKMASTPGVSATLFNALAQANINILAIAQGCSEYNVTVVLKR 727
Query: 538 EDSIK 542
EDSI+
Sbjct: 728 EDSIR 742
Score = 53.9 bits (128), Expect(2) = e-124
Identities = 25/30 (83%), Positives = 29/30 (96%)
Frame = +1
Query: 542 KALRAVHSRFYLSRTTIAMGIIGPGLIGST 571
KALRAVHSRFY S+TT+AMGIIGPGL+G+T
Sbjct: 739 KALRAVHSRFYNSQTTMAMGIIGPGLMGAT 828
Score = 39.3 bits (90), Expect = 0.007
Identities = 20/63 (31%), Positives = 36/63 (56%)
Frame = +2
Query: 485 IPNCSILAAVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVIKREDSIKAL 544
I N +++ G MA PG ++T+F+A+ + NV I+Q SE+++ + E +KA+
Sbjct: 326 IDNLALVNVEGTGMAGVPGTASTIFSAVKEVGANVIMISQASSEHSVCFAVP-EKEVKAV 502
Query: 545 RAV 547
V
Sbjct: 503 AEV 511
>BI308719 similar to PIR|T06242|T06 aspartate kinase (EC 2.7.2.4) /
homoserine dehydrogenase (EC 1.1.1.3) precursor -
soybean, partial (21%)
Length = 591
Score = 295 bits (756), Expect(2) = 2e-83
Identities = 140/160 (87%), Positives = 150/160 (93%)
Frame = +1
Query: 640 KWKELREERGEVANLEKFAQHVHGNNFIPNTALVDCTADSIIAGHYYEWLCKGIHVITPN 699
KW+E+REE+GEVA+LEKF QHVHGN+ IPNT LVDCTADS+IA HY +WL KGIHVITPN
Sbjct: 112 KWREIREEKGEVADLEKFVQHVHGNHVIPNTVLVDCTADSVIASHYDDWLRKGIHVITPN 291
Query: 700 KKANSGPLEQYLRLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLETGDKILQIEGIFSG 759
KKANSGPL +YLRLRALQRQSYTHYFYEATVGAGLPI+ TLRGLLETGDKILQIEGIFSG
Sbjct: 292 KKANSGPLSEYLRLRALQRQSYTHYFYEATVGAGLPIIGTLRGLLETGDKILQIEGIFSG 471
Query: 760 TLSYIFNNFKDGRAFSEVVGEAKEAGYTEPDPRDDLSGTD 799
TLSYIFNNFKDG+ FSEVV EAKEAGYTEPDPRDDLSGTD
Sbjct: 472 TLSYIFNNFKDGQVFSEVVAEAKEAGYTEPDPRDDLSGTD 591
Score = 33.1 bits (74), Expect(2) = 2e-83
Identities = 16/24 (66%), Positives = 17/24 (70%)
Frame = +2
Query: 611 RLQF*KKNLTLICV*WA*LAQSQC 634
R Q *KKN TLICV*W QS+C
Sbjct: 11 RQQL*KKNQTLICV*WGSWVQSRC 82
>TC85113 homologue to PIR|T06242|T06242 aspartate kinase (EC 2.7.2.4) /
homoserine dehydrogenase (EC 1.1.1.3) precursor -
soybean, partial (14%)
Length = 414
Score = 119 bits (297), Expect(2) = 7e-52
Identities = 62/67 (92%), Positives = 63/67 (93%)
Frame = +1
Query: 477 GRLSQVAVIPNCSILAAVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVIK 536
G LSQVAVIPNCSILAAVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNIT VIK
Sbjct: 163 GSLSQVAVIPNCSILAAVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITDVIK 342
Query: 537 REDSIKA 543
EDS K+
Sbjct: 343 PEDSYKS 363
Score = 104 bits (260), Expect(2) = 7e-52
Identities = 54/55 (98%), Positives = 55/55 (99%)
Frame = +3
Query: 424 ASAIFAAVKDVGANVIMISQASSEHSVCFAVPEKEVKAVAEALQSRFRQALDNGR 478
ASAIFAAVKDVGANVIMISQASSEHSVCFAVPEKEVKAVAEALQSRFRQALDNG+
Sbjct: 3 ASAIFAAVKDVGANVIMISQASSEHSVCFAVPEKEVKAVAEALQSRFRQALDNGQ 167
Score = 38.5 bits (88), Expect = 0.012
Identities = 18/28 (64%), Positives = 23/28 (81%)
Frame = +2
Query: 531 ITVVIKREDSIKALRAVHSRFYLSRTTI 558
+ +++ + IKALRAVHSRFYLSRTTI
Sbjct: 326 LLMLLSQRIVIKALRAVHSRFYLSRTTI 409
>TC77477 homologue to GP|5305740|gb|AAD41796.1| precursor monofunctional
aspartokinase {Glycine max}, partial (84%)
Length = 2339
Score = 179 bits (454), Expect = 4e-45
Identities = 125/472 (26%), Positives = 241/472 (50%), Gaps = 8/472 (1%)
Frame = +3
Query: 89 VHKFGGTCMGSSQRIKNVGDIVLNDDSERKLVVVSAMSKVTDMMYDLINKAQS---RDES 145
V KFGG+ + S++R+ V +V++ ER +VV+SAM K T+ + KA S +
Sbjct: 420 VMKFGGSSVASAERMMEVAGLVMSFPEERPIVVLSAMGKTTNKLLLAGEKAVSCGVTNVC 599
Query: 146 YISSLDAVLEKHSATAHDILDGETLAIFLSKLHEDISNLKAMLRAIYIAGHVTESFTDFV 205
I L + + H T + L + S + + + L+ +L I + +T+ D++
Sbjct: 600 GIEELSFIKDLHLRTV------DQLGVDRSIIEKHLEALEQLLNGIAMMKELTKRTQDYL 761
Query: 206 VGHGELWSAQMLSLVIRKNGIDCKWMDTREVLIVNP---TSSNQVDPDYLESERRLEKWY 262
V GE S ++ + + K G+ + D E+ + T+++ ++ Y +RL +
Sbjct: 762 VSFGECMSTRIFAAYLNKLGVKARQYDAFEIGFITTDDFTNADILEATYPAVAKRLHGDW 941
Query: 263 SLNPCKVIIATGFIASTPENIP-TTLKRDGSDFSAAIMGSLFRARQVTIWTDVDGVYSAD 321
+P + I TGF+ ++ TTL R GSD +A +G ++ +W DVDGV + D
Sbjct: 942 LADPA-IAIVTGFLGKARKSCAVTTLGRGGSDLTATTIGKALGLPEIQVWKDVDGVLTCD 1118
Query: 322 PRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMRYGIPILIRNIFNLSAPGTKIC 381
P +A + L++ EA E++YFGA VLHP+++ P IP+ ++N +N +APGT I
Sbjct: 1119PNIYPKAEPVPFLTFDEAAELAYFGAQVLHPQSMRPAREGDIPVRVKNSYNPNAPGTLIT 1298
Query: 382 HPVVSDYEDKSNLQNYVKGFATIDNLALVNVEGTGMAGVPGTASAIFAAVKDVGANVIMI 441
+++ + + N+ ++++ T M G G + +F+ +D+G +V ++
Sbjct: 1299-------KERDMSKAVLTSIVLKRNVTMLDIVSTRMLGQYGFLAKVFSIFEDLGISVDVV 1457
Query: 442 SQASSEHSVCFAVPEKEVKAVAEALQSRFRQALDN-GRLSQVAVIPNCSILAAVGQKMAS 500
A+SE SV + ++ + E +Q ++ +++ V ++ N SI++ +G S
Sbjct: 1458--ATSEVSVSLTLDPSKLWS-RELIQQELDHVVEELEKIAVVNLLQNRSIISLIGNVQQS 1628
Query: 501 TPGVSATLFNALAKANINVRAIAQGCSEYNITVVIKREDSIKALRAVHSRFY 552
+ + F L + V+ I+QG S+ NI++V+ ++ + +RA+H F+
Sbjct: 1629SL-ILEKAFRVLRTLGVTVQMISQGASKVNISLVVNDSEAEECVRALHHAFF 1781
>BF632875 similar to PIR|T06242|T062 aspartate kinase (EC 2.7.2.4) /
homoserine dehydrogenase (EC 1.1.1.3) precursor -
soybean, partial (2%)
Length = 355
Score = 177 bits (449), Expect = 2e-44
Identities = 90/97 (92%), Positives = 90/97 (92%)
Frame = +1
Query: 1 MASSLSSSLSHFSRISVTSLQHDYNNKIPADSQCRHFLLSRRFHSLRKGITLPRRRESPS 60
MASSLSSSLSHFSRISVTSLQHDYNNKIPADSQCRHFLLSRRFHSLRKGITLPRRRESPS
Sbjct: 85 MASSLSSSLSHFSRISVTSLQHDYNNKIPADSQCRHFLLSRRFHSLRKGITLPRRRESPS 264
Query: 61 SGICASLTDVSVNVAVEEKELSKGDSWSVHKFGGTCM 97
SGICASLT VEEKELSKGDSWSVHKFGGTCM
Sbjct: 265 SGICASLT-------VEEKELSKGDSWSVHKFGGTCM 354
>BE248159 homologue to PIR|T06246|T062 aspartate kinase (EC 2.7.2.4) /
homoserine dehydrogenase (EC 1.1.1.3) precursor -
soybean (fragment), partial (8%)
Length = 237
Score = 164 bits (415), Expect = 1e-40
Identities = 79/79 (100%), Positives = 79/79 (100%)
Frame = +1
Query: 321 DPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMRYGIPILIRNIFNLSAPGTKI 380
DPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMRYGIPILIRNIFNLSAPGTKI
Sbjct: 1 DPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMRYGIPILIRNIFNLSAPGTKI 180
Query: 381 CHPVVSDYEDKSNLQNYVK 399
CHPVVSDYEDKSNLQNYVK
Sbjct: 181 CHPVVSDYEDKSNLQNYVK 237
>TC87100 homologue to GP|9293891|dbj|BAB01794.1 contains similarity to
uridylate kinase~gene_id:MVE11.4 {Arabidopsis thaliana},
partial (58%)
Length = 891
Score = 35.8 bits (81), Expect = 0.079
Identities = 18/47 (38%), Positives = 30/47 (63%)
Frame = +2
Query: 292 SDFSAAIMGSLFRARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQE 338
+D +AA+ + A V T+VDGV+ DP++ +A +L TL+YQ+
Sbjct: 710 TDTAAALRCAEINAEVVLKATNVDGVFDDDPKRNPQARLLDTLTYQD 850
>TC81087 homologue to GP|530207|gb|AAA66338.1|| heat shock protein {Glycine
max}, partial (41%)
Length = 1318
Score = 32.0 bits (71), Expect = 1.1
Identities = 40/192 (20%), Positives = 76/192 (38%), Gaps = 12/192 (6%)
Frame = +2
Query: 755 GIFSGTLSYIFNNFKDGRAFSEVVGEAKEAGYTEPDPRDDLSGTDVARKVIILARESGLK 814
GIF +S + + RA V+ +A + ++ P D++ G+ K I A+ +
Sbjct: 311 GIFFQAISNVAGE-ESARAVERVLKQALKKLPSQSPPPDEVPGSTALIKAIRRAQAAQKS 487
Query: 815 LELSNIPIESLVPEPLR----------ACASAQEFMQQLPKFDQEFAKKQEDA--DNAGE 862
+++ ++ L+ L A + ++ K + KK E A D +
Sbjct: 488 RGDTHLAVDQLILGILEDSQIGDLFKEAGVAVSRVKTEVEKLRGKDGKKVESASGDTNFQ 667
Query: 863 VLRYVGVVDVTNKKGVVELRKYKKDHPFAQLSGSDNIIAFTTRRYKNQPLIVRGPGAGAQ 922
L+ G D+ + G K D + ++ +RR KN P+++ PG G
Sbjct: 668 ALKTYGR-DLVEQAG-------KLDPVIGRDEEIRRVVRILSRRTKNNPVLIGEPGVGKT 823
Query: 923 VTAGGIFSDILR 934
G+ I+R
Sbjct: 824 AVVEGLAQRIVR 859
>TC87630 similar to GP|11385453|gb|AAG34809.1 glutathione S-transferase GST
19 {Glycine max}, partial (98%)
Length = 967
Score = 31.2 bits (69), Expect = 2.0
Identities = 18/54 (33%), Positives = 27/54 (49%), Gaps = 3/54 (5%)
Frame = +2
Query: 638 MSKWKELREERGEVA--NLEKFAQ-HVHGNNFIPNTALVDCTADSIIAGHYYEW 688
+S W + EE GE+ N EKF H G+NFI + + DC + Y+ +
Sbjct: 551 ISYWLNVLEELGEMELINAEKFPSLHEWGHNFIQTSPIKDCIPPKEMVVEYFSF 712
>TC77222 weakly similar to GP|19911209|dbj|BAB86931. glucosyltransferase-13
{Vigna angularis}, partial (76%)
Length = 1643
Score = 30.4 bits (67), Expect = 3.3
Identities = 16/43 (37%), Positives = 23/43 (53%), Gaps = 5/43 (11%)
Frame = -3
Query: 370 IFNLSAPGTKICH-----PVVSDYEDKSNLQNYVKGFATIDNL 407
IF+ S P K+ H V ++ K QN+V GF+TI N+
Sbjct: 915 IFDHSIPNQKLSH*FVFEKVFHHFQKKHTKQNFVVGFSTISNI 787
>TC83315 weakly similar to GP|19911209|dbj|BAB86931. glucosyltransferase-13
{Vigna angularis}, partial (42%)
Length = 1048
Score = 30.4 bits (67), Expect = 3.3
Identities = 16/43 (37%), Positives = 24/43 (55%), Gaps = 5/43 (11%)
Frame = -3
Query: 370 IFNLSAPGTKICHPVVSD-----YEDKSNLQNYVKGFATIDNL 407
IF+ S P K+ H V + ++ K QN+V GF+TI N+
Sbjct: 869 IFDHSIPNQKLVHLFVFEKVFHHFQKKHTKQNFVVGFSTISNI 741
>TC79349 similar to PIR|S65668|S65668 preprotein translocase secA precursor
- garden pea, partial (19%)
Length = 857
Score = 30.0 bits (66), Expect = 4.3
Identities = 42/162 (25%), Positives = 65/162 (39%)
Frame = +3
Query: 2 ASSLSSSLSHFSRISVTSLQHDYNNKIPADSQCRHFLLSRRFHSLRKGITLPRRRESPSS 61
ASSL SS + + S L H +P S LSR FH ++ RR S S
Sbjct: 240 ASSLCSSFTSTTSHSQHRLHHRKTLNLPGSS-----FLSREFHLNSASVSKTRRSRSRRS 404
Query: 62 GICASLTDVSVNVAVEEKELSKGDSWSVHKFGGTCMGSSQRIKNVGDIVLNDDSERKLVV 121
G +VAV G F G G + R + + + + E +
Sbjct: 405 G----------SVAVASLGGLLGGI-----FKGNDTGEATRKQYAATVNVINGLEANISK 539
Query: 122 VSAMSKVTDMMYDLINKAQSRDESYISSLDAVLEKHSATAHD 163
+S S++ D ++L +AQ R+ SLD++L + A +
Sbjct: 540 LSD-SELRDKTFELRERAQKRE-----SLDSLLPEAFAVVRE 647
>TC78184 similar to GP|17979207|gb|AAL49842.1 unknown protein {Arabidopsis
thaliana}, partial (76%)
Length = 1897
Score = 29.6 bits (65), Expect = 5.7
Identities = 24/88 (27%), Positives = 41/88 (46%), Gaps = 2/88 (2%)
Frame = +3
Query: 9 LSHFSRISVTSLQHDYNNKIPADSQCRHFLLSRRFHSLRKGITLPRRRESPSSGICASLT 68
+ HF RI + L +N + R ++SRRFHSL + E+ + ++
Sbjct: 150 IDHFDRIPDSLLLLVFNKIGDVKALGRCCVVSRRFHSLVPQV------ENVVVRVDCVIS 311
Query: 69 DVSVNVAVEEKELSKGDSWSVHK--FGG 94
D N +V + S+G W++ + FGG
Sbjct: 312 DDDSNSSVNSSDKSRGPFWNLLRLVFGG 395
>AW691925
Length = 649
Score = 29.6 bits (65), Expect = 5.7
Identities = 42/162 (25%), Positives = 67/162 (40%), Gaps = 5/162 (3%)
Frame = +2
Query: 32 SQCRHFLLSRRFHSLRKGITLPRRRESPSSGICASLTDVSVNVAVEEKELSKGDSWSVHK 91
S+C HF L +H L L RRR+ + +LS G W +H
Sbjct: 263 SRCTHFCLGYTYHGLSD--FLSRRRQ------------------IRHLDLS-GAPWYIH- 376
Query: 92 FGGTCMGSSQRIKNVGDIVLNDDSERKLVVVSAMSKVTD-MMYDLINKAQSRDESYISSL 150
++I+ + D+V + S + +++ SK+TD +Y LI +S E Y+
Sbjct: 377 --------DEQIETLADLVTDIIS----INLTSCSKLTDYAVYILITTCRSLSELYMG-- 514
Query: 151 DAVLEKHSATAHDIL----DGETLAIFLSKLHEDISNLKAML 188
D + + S T + L D T LS H+ N + ML
Sbjct: 515 DTKVGEGSETFPEQLLLCPDNNTRLQSLSLSHQKFLNGEVML 640
>TC79978 weakly similar to GP|20197257|gb|AAC28988.2 putative ABC
transporter; alternative splicing isoform gene
prediction data combined with cDNA, partial (7%)
Length = 832
Score = 29.3 bits (64), Expect = 7.4
Identities = 12/27 (44%), Positives = 18/27 (66%)
Frame = -1
Query: 22 HDYNNKIPADSQCRHFLLSRRFHSLRK 48
+DYN P Q ++F+ + RFH+LRK
Sbjct: 349 NDYNKGRPTKPQHQYFMTACRFHNLRK 269
>TC78118 similar to PIR|T06528|T06528 lectin - garden pea, partial (77%)
Length = 1110
Score = 29.3 bits (64), Expect = 7.4
Identities = 19/64 (29%), Positives = 31/64 (47%), Gaps = 1/64 (1%)
Frame = +1
Query: 371 FNLSAPGT-KICHPVVSDYEDKSNLQNYVKGFATIDNLALVNVEGTGMAGVPGTASAIFA 429
+ + P T +I P VS ++ N+ ++V F+ + +EGTG GVP F
Sbjct: 265 YGTTIPSTGRILTPPVSLWDTAGNVASFVTSFSFL-------IEGTGGYGVPTDGLVFFI 423
Query: 430 AVKD 433
A +D
Sbjct: 424 APQD 435
>TC89482 similar to SP|Q9M3H5|AHM1_ARATH Potential cadmium/zinc-transporting
ATPase HMA1 (EC 3.6.3.3) (EC 3.6.3.5). [Mouse-ear
cress], partial (41%)
Length = 1100
Score = 28.9 bits (63), Expect = 9.7
Identities = 21/70 (30%), Positives = 33/70 (47%)
Frame = +3
Query: 398 VKGFATIDNLALVNVEGTGMAGVPGTASAIFAAVKDVGANVIMISQASSEHSVCFAVPEK 457
+KG +D LA + G T +F A++ V + I ++ S+ S C EK
Sbjct: 114 LKGGHVLDALASCHTIAFDKTGTLTTGGLVFKAIEPVYGHHIR-NKESNISSCCVPTCEK 290
Query: 458 EVKAVAEALQ 467
E AVA A++
Sbjct: 291 EALAVAAAME 320
>BG454676
Length = 666
Score = 28.9 bits (63), Expect = 9.7
Identities = 12/34 (35%), Positives = 16/34 (46%)
Frame = +3
Query: 678 DSIIAGHYYEWLCKGIHVITPNKKANSGPLEQYL 711
D+I H+Y WLC+ +H K N P L
Sbjct: 9 DAIYPPHFYSWLCRRVHFKNT*SKRN*RPCTDIL 110
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.135 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,080,499
Number of Sequences: 36976
Number of extensions: 346152
Number of successful extensions: 1552
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 1530
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1550
length of query: 942
length of database: 9,014,727
effective HSP length: 105
effective length of query: 837
effective length of database: 5,132,247
effective search space: 4295690739
effective search space used: 4295690739
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146585.12


