

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146583.2 + phase: 0 /pseudo
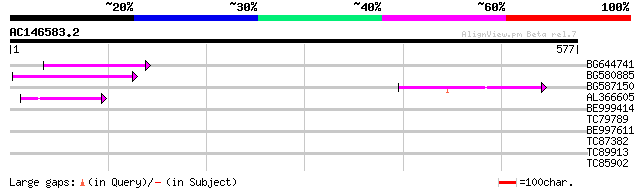
(577 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG644741 85 7e-17
BG580885 62 6e-10
BG587150 similar to GP|13592175|gb ppg3 {Leishmania major}, part... 55 1e-07
AL366605 45 8e-05
BE999414 37 0.016
TC79789 similar to PIR|D96519|D96519 mysoin-like protein 11013-... 34 0.13
BE997611 31 1.5
TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulch... 31 1.5
TC89913 similar to PIR|H96569|H96569 unknown protein 54928-5675... 30 1.9
TC85902 similar to SP|Q9XF97|RL4_PRUAR 60S ribosomal protein L4 ... 29 5.7
>BG644741
Length = 735
Score = 85.1 bits (209), Expect = 7e-17
Identities = 40/109 (36%), Positives = 58/109 (52%)
Frame = -2
Query: 35 LHLFPFSLRDRASAWFHSLEVGSITSWDDMRRAFLARFFPPSKTAKLRDQITRFNQKDGE 94
L +FP SL A WF L SI +W+ +R FLAR++P SK D++ F GE
Sbjct: 587 LRVFPLSLMGEADIWFTELPYNSIFTWNQLRDVFLARYYPVSKKLNHNDRVNNFVALPGE 408
Query: 95 SLYEAWEHFKEMLRLCPHHGLEKWLIVHTFYNGLSDTTKMSVDAAAGGA 143
S+ +W+ F LR P+H ++ + FY G D K+ +D AGG+
Sbjct: 407 SVSSSWDRFTSFLRSVPNHRIDDDSLKEYFYRGQDDNNKVVLDTIAGGS 261
>BG580885
Length = 609
Score = 62.0 bits (149), Expect = 6e-10
Identities = 34/129 (26%), Positives = 68/129 (52%), Gaps = 2/129 (1%)
Frame = +3
Query: 4 FSGSPTEDPNLHISSFLRLSGTIKENQEAVRLHLFPFSLRDRASAWFH-SLEVGSIT-SW 61
F G+P E P H+S F ++ + ++ +FP +L + ++ W+ ++E I+ SW
Sbjct: 54 FRGTPNESPITHLSRFNKVCRANNASSVEMQKKIFPVTLEEESALWYDLNIEPYYISLSW 233
Query: 62 DDMRRAFLARFFPPSKTAKLRDQITRFNQKDGESLYEAWEHFKEMLRLCPHHGLEKWLIV 121
D+++ +FL ++ +LR ++ +Q + E + + + +L+ P HGLE +I
Sbjct: 234 DEIKLSFLQAYYEIEPVEELRSELMGIHQGEKERVRSYFLRLQWILKRWPEHGLEDDVIK 413
Query: 122 HTFYNGLSD 130
F NGL +
Sbjct: 414 GVFVNGLRE 440
>BG587150 similar to GP|13592175|gb ppg3 {Leishmania major}, partial (1%)
Length = 754
Score = 54.7 bits (130), Expect = 1e-07
Identities = 42/160 (26%), Positives = 74/160 (46%), Gaps = 9/160 (5%)
Frame = +2
Query: 396 FGEALSRMPLYTKFLKEIFSKKKAMDHKETIALTRESSAIVKKQPPKLR---------DP 446
F + + +PL F + F+ + M +ET + + + K++ D
Sbjct: 167 FQKRVFMIPLEKPFEEAYFTHRLWMFFRETRETEEDIRRMFCEAREKMKKRITLKKKSDS 346
Query: 447 GSFVIPCVIGKETVDKALCDLGASVSLLPLSLFKRMGIGELKPTEMILKLADRSTIPLVG 506
G F I + ALC+ GASV +LP + +G+ ++ P++ D S G
Sbjct: 347 GKFAISWTMKGIEFPHALCNTGASVIILPRDMADHLGL-KVDPSKESFTFVDCSQRSSGG 523
Query: 507 YIEDIPVKIEGIYIPTDFVVVDIEEDHDVPIILGRPFLAT 546
I D+ V+I +P DF V+DI+ + + ++L R FL+T
Sbjct: 524 IIRDLEVQIGNALVPVDFHVLDIKINWNSSLLLRRAFLST 643
>AL366605
Length = 422
Score = 45.1 bits (105), Expect = 8e-05
Identities = 25/87 (28%), Positives = 46/87 (52%)
Frame = -3
Query: 12 PNLHISSFLRLSGTIKENQEAVRLHLFPFSLRDRASAWFHSLEVGSITSWDDMRRAFLAR 71
P HI ++R G K+N +++ +H F SL + A+ W+ SL I ++D++ AF +
Sbjct: 291 PQNHIIKYVRKMGNYKDN-DSLMIHCFQDSLMEDAAEWYTSLSKNDIHTFDELAAAFKSH 115
Query: 72 FFPPSKTAKLRDQITRFNQKDGESLYE 98
+ ++ R+ + +QK ES E
Sbjct: 114 YGFNTRLKPNREFLRSLSQKKEESFRE 34
>BE999414
Length = 613
Score = 37.4 bits (85), Expect = 0.016
Identities = 22/64 (34%), Positives = 33/64 (51%)
Frame = +3
Query: 9 TEDPNLHISSFLRLSGTIKENQEAVRLHLFPFSLRDRASAWFHSLEVGSITSWDDMRRAF 68
T DP+ H+ + + + T + + AV+ LF +LR A WF +L SI SW D+ F
Sbjct: 27 T*DPDEHMEN-IEVVLTYRSVRGAVKCKLFVTTLRRGAMTWFKNLRRNSIGSWGDLCHEF 203
Query: 69 LARF 72
F
Sbjct: 204TTHF 215
>TC79789 similar to PIR|D96519|D96519 mysoin-like protein 11013-7318
[imported] - Arabidopsis thaliana, partial (21%)
Length = 1097
Score = 34.3 bits (77), Expect = 0.13
Identities = 44/185 (23%), Positives = 79/185 (41%), Gaps = 5/185 (2%)
Frame = +3
Query: 210 RPNQNFYKNPQGSYGQTAPPGYTNNQRVAQKSSLEILLENCMMNQNKQLQELKNQTGSLN 269
+PNQ+ YK P +Y Q + Y++ LE+ + ++ Q L+++ LN
Sbjct: 312 QPNQDVYKKP--NYVQISVESYSHLTG----------LEDQVKTYEEKAQTLEDEINELN 455
Query: 270 DSLSKLNTKVDS----IATHTKMLETQISQVAQQVAISS*TPGVFPGQTETNPKAHVNAI 325
+ LS NT++++ + H K+ E +S + A + T + A A
Sbjct: 456 EKLSAANTEINTKEALVKQHAKVAEEAVSGWEKAEAEALALKNSLESVTLSKLTAEDQAS 635
Query: 326 SLGGNKLEDTITKAKSVKGESVKLLGEKDAIKT-LLDKNKALNPLRLTKLNLEAQFAKFL 384
L G L++ + + +++K E + + KT LDK K +LEA+ F
Sbjct: 636 QLDG-ALKECMRQIRNLKEEHELKIQDISLAKTKQLDKIKG---------DLEARIRNFE 785
Query: 385 NILKR 389
L R
Sbjct: 786 QELLR 800
>BE997611
Length = 547
Score = 30.8 bits (68), Expect = 1.5
Identities = 15/53 (28%), Positives = 28/53 (52%)
Frame = -2
Query: 379 QFAKFLNILKRICIKIPFGEALSRMPLYTKFLKEIFSKKKAMDHKETIALTRE 431
+F +ILK + +P E + ++P KFL+E+ + +E I+L+ E
Sbjct: 426 KFNALHHILKGYKVNMPITETVEQIPCCIKFLQELLKTNANLSEEEFISLSSE 268
>TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulchellus},
partial (7%)
Length = 2304
Score = 30.8 bits (68), Expect = 1.5
Identities = 21/81 (25%), Positives = 34/81 (41%), Gaps = 13/81 (16%)
Frame = +2
Query: 8 PTEDPNLHISSFLRLSGTI------KENQEAVRLHLFPFSLRDRASAWFHSLE------- 54
P + NL + FL TI KE E ++ + L+ A W+ +L+
Sbjct: 629 PDFEGNLQLDDFLDWLQTIERVFEYKEVPEEQKVKIVAAKLKKHALIWWENLKRRRKREG 808
Query: 55 VGSITSWDDMRRAFLARFFPP 75
I +WD MR+ ++ PP
Sbjct: 809 KSKIKTWDKMRQKLTRKYLPP 871
>TC89913 similar to PIR|H96569|H96569 unknown protein 54928-56750
[imported] - Arabidopsis thaliana, partial (37%)
Length = 761
Score = 30.4 bits (67), Expect = 1.9
Identities = 21/88 (23%), Positives = 40/88 (44%), Gaps = 3/88 (3%)
Frame = +2
Query: 364 KALNPL---RLTKLNLEAQFAKFLNILKRICIKIPFGEALSRMPLYTKFLKEIFSKKKAM 420
K +NPL K NL +++ IC +PF + + +PL++ +KK+
Sbjct: 44 KHINPLINFARKKQNLSLNSLMAMSVASSICTLLPFSKPHNSVPLFSS-----QTKKRCR 208
Query: 421 DHKETIALTRESSAIVKKQPPKLRDPGS 448
++ R+++++V K L D S
Sbjct: 209 IIISSVTEDRQAASVVSKDKKVLEDSSS 292
>TC85902 similar to SP|Q9XF97|RL4_PRUAR 60S ribosomal protein L4 (L1).
[Apricot] {Prunus armeniaca}, complete
Length = 1571
Score = 28.9 bits (63), Expect = 5.7
Identities = 25/100 (25%), Positives = 44/100 (44%)
Frame = +1
Query: 336 ITKAKSVKGESVKLLGEKDAIKTLLDKNKALNPLRLTKLNLEAQFAKFLNILKRICIKIP 395
+ +AK V + +++ + + K + L K L K LN++ R+ P
Sbjct: 931 LPRAKMVNSDLTRIINSDEVQSVVRPIKKDVKRATLKKNPL-----KNLNVMLRLN---P 1086
Query: 396 FGEALSRMPLYTKFLKEIFSKKKAMDHKETIALTRESSAI 435
+ + RM L + + + SKK+ D K I E+SAI
Sbjct: 1087 YAKTAKRMALLAE-AERVKSKKEKFDKKRKIVSKEEASAI 1203
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.134 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,789,385
Number of Sequences: 36976
Number of extensions: 206599
Number of successful extensions: 1020
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 1011
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1019
length of query: 577
length of database: 9,014,727
effective HSP length: 101
effective length of query: 476
effective length of database: 5,280,151
effective search space: 2513351876
effective search space used: 2513351876
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146583.2


