

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146574.14 - phase: 0
(211 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
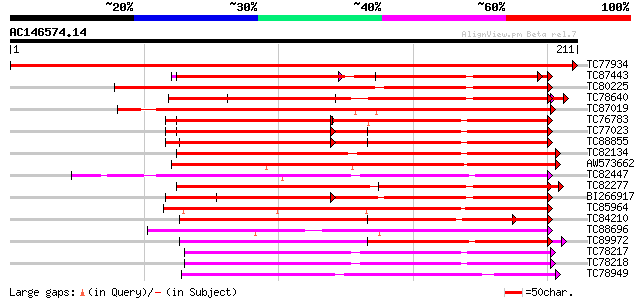
Score E
Sequences producing significant alignments: (bits) Value
TC77934 similar to GP|9965747|gb|AAG10150.1| calmodulin-like MSS... 422 e-119
TC87443 similar to GP|13397927|emb|CAC34625. putative calmodulin... 119 1e-27
TC80225 similar to GP|14589311|emb|CAC43238. calcium binding pro... 113 5e-26
TC78640 homologue to GP|13397927|emb|CAC34625. putative calmodul... 111 2e-25
TC87019 weakly similar to GP|12003380|gb|AAG43547.1 Avr9/Cf-9 ra... 107 3e-24
TC76783 calmodulin 1 [Medicago truncatula] 107 3e-24
TC77023 calmodulin 2 [Medicago truncatula] 107 3e-24
TC88855 PIR|A30900|MCBH calmodulin - barley, complete 107 4e-24
TC82134 similar to PIR|T07949|T07949 calcium binding protein - l... 103 5e-23
AW573662 weakly similar to PIR|D96689|D966 calmodulin-related pr... 102 1e-22
TC82447 similar to GP|14589311|emb|CAC43238. calcium binding pro... 102 1e-22
TC82277 weakly similar to PIR|A84532|A84532 probable calmodulin-... 98 3e-21
BI266917 PIR|S01173|MC calmodulin [validated] - fruit fly (Droso... 97 6e-21
TC85964 similar to PIR|D84841|D84841 calmodulin-like protein [im... 96 1e-20
TC84210 homologue to PIR|T07122|T07122 calmodulin CAM5 - soybean... 88 2e-18
TC88696 weakly similar to PIR|T10620|T10620 probable calcium-bin... 87 4e-18
TC89972 similar to GP|22296346|dbj|BAC10116. putative caltractin... 86 1e-17
TC78217 weakly similar to GP|20161870|dbj|BAB90783. hypothetical... 85 2e-17
TC78218 weakly similar to GP|12963415|gb|AAK11255.1 regulator of... 84 3e-17
TC78949 similar to GP|16215471|emb|CAC82999. calcium-dependent p... 82 2e-16
>TC77934 similar to GP|9965747|gb|AAG10150.1| calmodulin-like MSS3
{Arabidopsis thaliana}, partial (80%)
Length = 1325
Score = 422 bits (1084), Expect = e-119
Identities = 211/211 (100%), Positives = 211/211 (100%)
Frame = +1
Query: 1 MPTILLRIFLLYNVVNSFLISLVPKKLITFFPHSWFTHQTLTTPSSTSKRGLVFTKTITM 60
MPTILLRIFLLYNVVNSFLISLVPKKLITFFPHSWFTHQTLTTPSSTSKRGLVFTKTITM
Sbjct: 97 MPTILLRIFLLYNVVNSFLISLVPKKLITFFPHSWFTHQTLTTPSSTSKRGLVFTKTITM 276
Query: 61 DPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEE 120
DPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEE
Sbjct: 277 DPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEE 456
Query: 121 FRELYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKK 180
FRELYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKK
Sbjct: 457 FRELYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKK 636
Query: 181 MIGTVDVDGNGLVDYKEFKQMMKGGGFTALS 211
MIGTVDVDGNGLVDYKEFKQMMKGGGFTALS
Sbjct: 637 MIGTVDVDGNGLVDYKEFKQMMKGGGFTALS 729
>TC87443 similar to GP|13397927|emb|CAC34625. putative calmodulin-related
protein {Medicago sativa}, partial (69%)
Length = 918
Score = 119 bits (297), Expect = 1e-27
Identities = 57/140 (40%), Positives = 99/140 (70%)
Frame = +3
Query: 63 NELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFR 122
+E++++F FD+N DG+I++ EL + L LG +E+ +M+E++D N DG +D++EF
Sbjct: 24 DEVRKIFSKFDKNGDGKISRSELKEMLLTLGSETTSEEVKRMMEELDQNGDGFIDLKEFA 203
Query: 123 ELYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMI 182
+ + +E ++E ++R+AF+++D + +G IS +EL +VL+ LG K ++ DCKKMI
Sbjct: 204 DFH---CTEPGKDESSELRDAFDLYDLDKNGLISANELHAVLMKLGEK--CSLNDCKKMI 368
Query: 183 GTVDVDGNGLVDYKEFKQMM 202
VDVDG+G V+++EFK+MM
Sbjct: 369 SNVDVDGDGNVNFEEFKKMM 428
Score = 55.1 bits (131), Expect = 2e-08
Identities = 26/62 (41%), Positives = 45/62 (71%)
Frame = +3
Query: 137 EEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLVDYK 196
++++R+ F+ FD+NGDG IS EL+ +L++LG + T E+ K+M+ +D +G+G +D K
Sbjct: 21 DDEVRKIFSKFDKNGDGKISRSELKEMLLTLGSE--TTSEEVKRMMEELDQNGDGFIDLK 194
Query: 197 EF 198
EF
Sbjct: 195 EF 200
Score = 42.7 bits (99), Expect = 1e-04
Identities = 21/64 (32%), Positives = 38/64 (58%)
Frame = +3
Query: 61 DPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEE 120
+ +EL+ F ++D + +G I+ EL+ L LG + +MI +DV+ DG V+ EE
Sbjct: 234 ESSELRDAFDLYDLDKNGLISANELHAVLMKLGEKCSLNDCKKMISNVDVDGDGNVNFEE 413
Query: 121 FREL 124
F+++
Sbjct: 414 FKKM 425
>TC80225 similar to GP|14589311|emb|CAC43238. calcium binding protein
{Sesbania rostrata}, partial (87%)
Length = 704
Score = 113 bits (283), Expect = 5e-26
Identities = 64/163 (39%), Positives = 101/163 (61%)
Frame = +3
Query: 40 TLTTPSSTSKRGLVFTKTITMDPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDK 99
T+ T +T + D +ELK VF FD N DG+I+ EL++ L +LG +P
Sbjct: 12 TMATNPNTETTTTTKSSVYLGDMDELKTVFTRFDTNGDGKISVTELDNILRSLGSTVPKD 191
Query: 100 ELSQMIEKIDVNRDGCVDIEEFRELYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDE 159
EL +++E +D +RDG +++ EF S ++ D E +REAF+++D++ +G IS E
Sbjct: 192 ELQRVMEDLDTDRDGFINLAEFAAFCRSGSADGDVSE---LREAFDLYDKDKNGLISATE 362
Query: 160 LRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLVDYKEFKQMM 202
L VL +LG+K +VE+C MI +VD DG+G V+++EFK+MM
Sbjct: 363 LCQVLNTLGMK--CSVEECHTMIKSVDSDGDGNVNFEEFKKMM 485
>TC78640 homologue to GP|13397927|emb|CAC34625. putative calmodulin-related
protein {Medicago sativa}, partial (84%)
Length = 1003
Score = 111 bits (277), Expect = 2e-25
Identities = 56/143 (39%), Positives = 95/143 (66%)
Frame = +2
Query: 60 MDPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIE 119
MD E++++F FD+N DG+I++ EL + + LG +E+++M+E++D N DG +D++
Sbjct: 413 MDQEEVRKIFNKFDKNGDGKISRTELKEMMTALGSKTTTEEVTRMMEELDRNGDGYIDLK 592
Query: 120 EFRELYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCK 179
EF EL+ + +++REAF ++D + +G IS EL +V+ LG K ++ DC+
Sbjct: 593 EFGELHNG------GGDTKELREAFEMYDLDKNGLISAKELHAVMRRLGEK--CSLGDCR 748
Query: 180 KMIGTVDVDGNGLVDYKEFKQMM 202
KMIG VD D +G V+++EFK+MM
Sbjct: 749 KMIGNVDADADGNVNFEEFKKMM 817
Score = 65.1 bits (157), Expect = 2e-11
Identities = 35/87 (40%), Positives = 55/87 (62%)
Frame = +2
Query: 122 RELYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKM 181
+++ SI D+EE +R+ FN FD+NGDG IS EL+ ++ +LG K T E+ +M
Sbjct: 383 KDINSSISQAMDQEE---VRKIFNKFDKNGDGKISRTELKEMMTALGSKT--TTEEVTRM 547
Query: 182 IGTVDVDGNGLVDYKEFKQMMKGGGFT 208
+ +D +G+G +D KEF ++ GGG T
Sbjct: 548 MEELDRNGDGYIDLKEFGELHNGGGDT 628
Score = 49.7 bits (117), Expect = 8e-07
Identities = 32/122 (26%), Positives = 59/122 (48%)
Frame = +2
Query: 82 KKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFRELYESIMSERDEEEEEDMR 141
+K++N S+ + +E+ ++ K D N DG + E +E+ ++ S+ EE M
Sbjct: 380 QKDINSSISQA---MDQEEVRKIFNKFDKNGDGKISRTELKEMMTALGSKTTTEEVTRMM 550
Query: 142 EAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLVDYKEFKQM 201
E D+NGDG+I + E + G ++ ++ D+D NGL+ KE +
Sbjct: 551 EEL---DRNGDGYIDLKEFGELH-----NGGGDTKELREAFEMYDLDKNGLISAKELHAV 706
Query: 202 MK 203
M+
Sbjct: 707 MR 712
>TC87019 weakly similar to GP|12003380|gb|AAG43547.1 Avr9/Cf-9 rapidly
elicited protein 31 {Nicotiana tabacum}, partial (76%)
Length = 967
Score = 107 bits (267), Expect = 3e-24
Identities = 63/171 (36%), Positives = 105/171 (60%), Gaps = 8/171 (4%)
Frame = +3
Query: 41 LTTPSSTSKRGLVFTKTITMDPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKE 100
L++PS + + ++ +++ L+R+F MFD+N D IT +E++ +L LG+ KE
Sbjct: 216 LSSPSKSFR-----LRSQSLNSLRLRRIFDMFDKNGDSMITVEEISQALNLLGLEAEFKE 380
Query: 101 LSQMIEKIDVNRDGCVDIEEFRELYES-------IMSERDEE-EEEDMREAFNVFDQNGD 152
+ MI+ + + E+F L+ES + +E DEE + ED+ EAF VFD++GD
Sbjct: 381 VDSMIKSYIKPGNVGLTYEDFVGLHESLGDTYFSVAAETDEETQNEDLWEAFKVFDEDGD 560
Query: 153 GFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLVDYKEFKQMMK 203
G+IS EL+ VL LGL +G +++ ++MI +VD + +G VD+ EFK MM+
Sbjct: 561 GYISAKELQVVLGKLGLVEGNLIDNVQRMILSVDTNHDGRVDFHEFKDMMR 713
>TC76783 calmodulin 1 [Medicago truncatula]
Length = 957
Score = 107 bits (267), Expect = 3e-24
Identities = 59/140 (42%), Positives = 87/140 (62%)
Frame = +3
Query: 63 NELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFR 122
+E K F +FD++ DG IT KEL + +LG + EL MI ++D + +G +D EF
Sbjct: 150 SEFKEAFSLFDKDGDGCITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL 329
Query: 123 ELYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMI 182
L M + D EEE ++EAF VFD++ +GFIS ELR V+ +LG K T E+ +MI
Sbjct: 330 NLMARKMKDTDSEEE--LKEAFRVFDKDQNGFISAAELRHVMTNLGEK--LTDEEVDEMI 497
Query: 183 GTVDVDGNGLVDYKEFKQMM 202
DVDG+G ++Y+EF ++M
Sbjct: 498 READVDGDGQINYEEFVKVM 557
Score = 60.5 bits (145), Expect = 5e-10
Identities = 31/83 (37%), Positives = 54/83 (64%), Gaps = 1/83 (1%)
Frame = +3
Query: 121 FRELYESIMSER-DEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCK 179
+R ++ M+++ +++ + +EAF++FD++GDG I+ EL +V+ SLG Q T + +
Sbjct: 96 YRRRRKATMADQLTDDQISEFKEAFSLFDKDGDGCITTKELGTVMRSLG--QNPTEAELQ 269
Query: 180 KMIGTVDVDGNGLVDYKEFKQMM 202
MI VD DGNG +D+ EF +M
Sbjct: 270 DMINEVDADGNGTIDFPEFLNLM 338
Score = 52.4 bits (124), Expect = 1e-07
Identities = 25/63 (39%), Positives = 40/63 (62%)
Frame = +3
Query: 59 TMDPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDI 118
T ELK F++FD++ +G I+ EL + NLG + D+E+ +MI + DV+ DG ++
Sbjct: 357 TDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDEEVDEMIREADVDGDGQINY 536
Query: 119 EEF 121
EEF
Sbjct: 537 EEF 545
>TC77023 calmodulin 2 [Medicago truncatula]
Length = 999
Score = 107 bits (267), Expect = 3e-24
Identities = 59/140 (42%), Positives = 87/140 (62%)
Frame = +1
Query: 63 NELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFR 122
+E K F +FD++ DG IT KEL + +LG + EL MI ++D + +G +D EF
Sbjct: 115 SEFKEAFSLFDKDGDGCITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL 294
Query: 123 ELYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMI 182
L M + D EEE ++EAF VFD++ +GFIS ELR V+ +LG K T E+ +MI
Sbjct: 295 NLMARKMKDTDSEEE--LKEAFRVFDKDQNGFISAAELRHVMTNLGEK--LTDEEVDEMI 462
Query: 183 GTVDVDGNGLVDYKEFKQMM 202
DVDG+G ++Y+EF ++M
Sbjct: 463 READVDGDGQINYEEFVKVM 522
Score = 61.2 bits (147), Expect = 3e-10
Identities = 30/69 (43%), Positives = 46/69 (66%)
Frame = +1
Query: 134 EEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLV 193
+E+ + +EAF++FD++GDG I+ EL +V+ SLG Q T + + MI VD DGNG +
Sbjct: 103 DEQISEFKEAFSLFDKDGDGCITTKELGTVMRSLG--QNPTEAELQDMINEVDADGNGTI 276
Query: 194 DYKEFKQMM 202
D+ EF +M
Sbjct: 277 DFPEFLNLM 303
Score = 52.4 bits (124), Expect = 1e-07
Identities = 25/63 (39%), Positives = 40/63 (62%)
Frame = +1
Query: 59 TMDPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDI 118
T ELK F++FD++ +G I+ EL + NLG + D+E+ +MI + DV+ DG ++
Sbjct: 322 TDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDEEVDEMIREADVDGDGQINY 501
Query: 119 EEF 121
EEF
Sbjct: 502 EEF 510
>TC88855 PIR|A30900|MCBH calmodulin - barley, complete
Length = 889
Score = 107 bits (266), Expect = 4e-24
Identities = 59/139 (42%), Positives = 86/139 (61%)
Frame = +1
Query: 64 ELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFRE 123
E K F +FD++ DG IT KEL + +LG + EL MI ++D + +G +D EF
Sbjct: 109 EFKEAFSLFDKDGDGCITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLN 288
Query: 124 LYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIG 183
L M + D EEE ++EAF VFD++ +GFIS ELR V+ +LG K T E+ +MI
Sbjct: 289 LMARKMKDTDSEEE--LKEAFRVFDKDQNGFISAAELRHVMTNLGEK--LTDEEVDEMIR 456
Query: 184 TVDVDGNGLVDYKEFKQMM 202
DVDG+G ++Y+EF ++M
Sbjct: 457 EADVDGDGQINYEEFVKVM 513
Score = 59.7 bits (143), Expect = 8e-10
Identities = 29/69 (42%), Positives = 46/69 (66%)
Frame = +1
Query: 134 EEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLV 193
+++ + +EAF++FD++GDG I+ EL +V+ SLG Q T + + MI VD DGNG +
Sbjct: 94 DDQIAEFKEAFSLFDKDGDGCITTKELGTVMRSLG--QNPTEAELQDMINEVDADGNGTI 267
Query: 194 DYKEFKQMM 202
D+ EF +M
Sbjct: 268 DFPEFLNLM 294
Score = 52.4 bits (124), Expect = 1e-07
Identities = 25/63 (39%), Positives = 40/63 (62%)
Frame = +1
Query: 59 TMDPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDI 118
T ELK F++FD++ +G I+ EL + NLG + D+E+ +MI + DV+ DG ++
Sbjct: 313 TDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDEEVDEMIREADVDGDGQINY 492
Query: 119 EEF 121
EEF
Sbjct: 493 EEF 501
>TC82134 similar to PIR|T07949|T07949 calcium binding protein - loblolly
pine, partial (27%)
Length = 630
Score = 103 bits (257), Expect = 5e-23
Identities = 55/143 (38%), Positives = 87/143 (60%)
Frame = +1
Query: 63 NELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFR 122
+E+K VF+ FD N DG+I+ +E + ++L I D + + +D ++DG +D +EF
Sbjct: 64 DEMKWVFEKFDTNKDGKISLEEYKAAAKSLDKGIGDPDAVKAFNVMDSDKDGFIDFKEFM 243
Query: 123 ELYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMI 182
E++ E ++ +EE+++ AF VFD NGDG IS +EL + LG + ++ CKKM+
Sbjct: 244 EMFN---GENNKIKEEEIKSAFQVFDINGDGKISAEELSQIFKRLG--ESCSLSACKKMV 408
Query: 183 GTVDVDGNGLVDYKEFKQMMKGG 205
VD DG+GL+D EF MM G
Sbjct: 409 KGVDGDGHGLIDLNEFTTMMMNG 477
>AW573662 weakly similar to PIR|D96689|D966 calmodulin-related protein
72976-72503 [imported] - Arabidopsis thaliana, partial
(29%)
Length = 615
Score = 102 bits (254), Expect = 1e-22
Identities = 58/150 (38%), Positives = 93/150 (61%), Gaps = 5/150 (3%)
Frame = +1
Query: 61 DPNELKRVFQMFDRNDDGRITKKELNDSLENLGI--FIPDKELSQMIEKIDVNRDGCVDI 118
+ + +++F++ D N+DG+I+ EL++ L LG KE MI +D N DG VDI
Sbjct: 103 ESGQFRQIFKLIDTNNDGKISTTELSEVLSCLGYNKCTASKEAESMIRVLDFNGDGFVDI 282
Query: 119 EEFRELYE---SIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTV 175
+EF + + + ++ + +E + +AF VFD + +G IS ELR VLV+LG + ++
Sbjct: 283 DEFMFVMKEDGNFGKGKEHDHDEYLMDAFLVFDIDKNGLISPKELRRVLVNLGC-ENCSL 459
Query: 176 EDCKKMIGTVDVDGNGLVDYKEFKQMMKGG 205
DCK+MI VD +G+G VD++EF+ MMK G
Sbjct: 460 RDCKRMIKGVDKNGDGFVDFEEFRSMMKIG 549
>TC82447 similar to GP|14589311|emb|CAC43238. calcium binding protein
{Sesbania rostrata}, partial (79%)
Length = 660
Score = 102 bits (253), Expect = 1e-22
Identities = 63/180 (35%), Positives = 106/180 (58%), Gaps = 1/180 (0%)
Frame = +2
Query: 24 PKKLITFFPHSWFTHQTLTTPSSTSKRGLVFTKTITMDPNELKRVFQMFDRNDDGRITKK 83
P KL TF P + T++++ S + + D +ELK+VF FD N DG+I+
Sbjct: 77 PNKLQTFKPMA--TNESIPVESDPTPT----PSPLLGDMDELKKVFNNFDANGDGKISVN 238
Query: 84 ELNDSLENLGIFIPDKE-LSQMIEKIDVNRDGCVDIEEFRELYESIMSERDEEEEEDMRE 142
EL L L +P +E L +++E++ +RDG +++ EF S + + + +R+
Sbjct: 239 ELETVLRTLRSDVPQQEELRRVMEELSTDRDGFINLSEFAAFCRS---DTTDGGDSALRD 409
Query: 143 AFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLVDYKEFKQMM 202
AF+++D++ +G IS +EL L LG+K +VE+C+ MI +VD DG+G V++ EFK+MM
Sbjct: 410 AFDLYDKDKNGLISTEELHLALDRLGMKC--SVEECQDMIKSVDSDGDGSVNFDEFKKMM 583
>TC82277 weakly similar to PIR|A84532|A84532 probable calmodulin-like
protein [imported] - Arabidopsis thaliana, partial (91%)
Length = 780
Score = 97.8 bits (242), Expect = 3e-21
Identities = 52/140 (37%), Positives = 86/140 (61%)
Frame = +3
Query: 63 NELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFR 122
+E+K VF FD N DG+I+++E +L++LG+ E+ + +D++ DG ++ EEF
Sbjct: 171 DEMKMVFDKFDSNKDGKISQQEYKATLKSLGMEKSVNEVPNIFRVVDLDGDGFINFEEFM 350
Query: 123 ELYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMI 182
E + R D++ AF FD+NGDG IS +E++ +L L ++ ++EDC++M+
Sbjct: 351 EAQKKGGGIRSL----DIQTAFRTFDKNGDGKISAEEIKEMLWKL--EERCSLEDCRRMV 512
Query: 183 GTVDVDGNGLVDYKEFKQMM 202
VD DG+G+VD EF MM
Sbjct: 513 RAVDTDGDGMVDMNEFVAMM 572
Score = 53.9 bits (128), Expect = 4e-08
Identities = 25/69 (36%), Positives = 44/69 (63%)
Frame = +3
Query: 138 EDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLVDYKE 197
++M+ F+ FD N DG IS E ++ L SLG++ ++V + + VD+DG+G ++++E
Sbjct: 171 DEMKMVFDKFDSNKDGKISQQEYKATLKSLGME--KSVNEVPNIFRVVDLDGDGFINFEE 344
Query: 198 FKQMMKGGG 206
F + K GG
Sbjct: 345 FMEAQKKGG 371
>BI266917 PIR|S01173|MC calmodulin [validated] - fruit fly (Drosophila
melanogaster), complete
Length = 576
Score = 96.7 bits (239), Expect = 6e-21
Identities = 55/125 (44%), Positives = 77/125 (61%)
Frame = +2
Query: 78 GRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFRELYESIMSERDEEEE 137
G IT KEL + +LG + EL MI ++D + +G +D EF + M + D EEE
Sbjct: 86 GTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSEEE 265
Query: 138 EDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLVDYKE 197
+REAF VFD++G+GFIS ELR V+ +LG K T E+ +MI D+DG+G V+Y+E
Sbjct: 266 --IREAFRVFDKDGNGFISAAELRHVMTNLGEK--LTDEEVDEMIREADIDGDGQVNYEE 433
Query: 198 FKQMM 202
F MM
Sbjct: 434 FVTMM 448
Score = 50.1 bits (118), Expect = 6e-07
Identities = 23/63 (36%), Positives = 40/63 (62%)
Frame = +2
Query: 59 TMDPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDI 118
T E++ F++FD++ +G I+ EL + NLG + D+E+ +MI + D++ DG V+
Sbjct: 248 TDSEEEIREAFRVFDKDGNGFISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNY 427
Query: 119 EEF 121
EEF
Sbjct: 428 EEF 436
>TC85964 similar to PIR|D84841|D84841 calmodulin-like protein [imported] -
Arabidopsis thaliana, partial (69%)
Length = 825
Score = 95.5 bits (236), Expect = 1e-20
Identities = 57/148 (38%), Positives = 90/148 (60%), Gaps = 3/148 (2%)
Frame = +1
Query: 58 ITMDPN-ELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPD-KELSQMIEKIDVNRDGC 115
IT+D EL + F++ DR++DG ++++EL L LG P +E++ M+ ++D + GC
Sbjct: 259 ITVDVQCELAQAFRLIDRDNDGVVSREELEAVLTRLGARPPTPEEIALMLSEVDSDGKGC 438
Query: 116 VDIEEFRELYESIMSE-RDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRT 174
+ +E S S D EE++REAF VFD + DG IS +EL V ++G + T
Sbjct: 439 ISVETIMNRVGSGSSSGSDPNPEEELREAFEVFDTDRDGRISAEELLRVFRAIG-DERCT 615
Query: 175 VEDCKKMIGTVDVDGNGLVDYKEFKQMM 202
+E+CK+MI VD +G+G V ++EF MM
Sbjct: 616 LEECKRMIAGVDKNGDGFVCFQEFSLMM 699
>TC84210 homologue to PIR|T07122|T07122 calmodulin CAM5 - soybean, partial
(96%)
Length = 627
Score = 88.2 bits (217), Expect = 2e-18
Identities = 49/126 (38%), Positives = 78/126 (61%)
Frame = +1
Query: 64 ELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFRE 123
E+K F +FD++ DG IT +EL + +L ++EL +MI ++D + +G ++ EF
Sbjct: 16 EIKEAFCLFDKDGDGCITVEELATVIRSLDQNPTEEELQEMINEVDADGNGTIEFVEFLN 195
Query: 124 LYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIG 183
L M E D +E D++EAF VFD++ +G+IS ELR V+++LG K T E+ +MI
Sbjct: 196 LMAKKMKETDADE--DLKEAFKVFDKDQNGYISASELRHVMINLGEK--LTDEEVDQMIK 363
Query: 184 TVDVDG 189
D+DG
Sbjct: 364 EADLDG 381
Score = 60.8 bits (146), Expect = 4e-10
Identities = 30/69 (43%), Positives = 49/69 (70%)
Frame = +1
Query: 134 EEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLV 193
E++ +++EAF +FD++GDG I+V+EL +V+ S L Q T E+ ++MI VD DGNG +
Sbjct: 1 EDQIVEIKEAFCLFDKDGDGCITVEELATVIRS--LDQNPTEEELQEMINEVDADGNGTI 174
Query: 194 DYKEFKQMM 202
++ EF +M
Sbjct: 175 EFVEFLNLM 201
>TC88696 weakly similar to PIR|T10620|T10620 probable calcium-binding
protein F21C20.130 - Arabidopsis thaliana, partial (17%)
Length = 648
Score = 87.4 bits (215), Expect = 4e-18
Identities = 55/165 (33%), Positives = 83/165 (49%), Gaps = 14/165 (8%)
Frame = +1
Query: 52 LVFTKTITMDPNELKRVFQMFDRNDDGRITKKELNDSLE---NLGIFIPDKELSQMIEKI 108
++F + +L R+F+ D N DG ++ +ELN L+ N +EL ++EK
Sbjct: 31 MLFMNMSILSSTDLHRIFEKLDTNCDGFVSLEELNSVLQRICNTSSQFSLEELESLVEKK 210
Query: 109 DVNRDGCVDIEEFRELYESIMSERDEEE-----------EEDMREAFNVFDQNGDGFISV 157
+ D EF Y SI E++EE E D+ + F VFD +GDGFI+
Sbjct: 211 SL------DFNEFLFFYNSISKEKNEENRGGDEDENDELERDLVKTFKVFDLDGDGFITS 372
Query: 158 DELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLVDYKEFKQMM 202
EL VL LG + +DC+ MI D + +G +D++EFK MM
Sbjct: 373 QELECVLKRLGFLDESSGKDCRSMIRFYDTNLDGRLDFQEFKNMM 507
>TC89972 similar to GP|22296346|dbj|BAC10116. putative caltractin {Oryza
sativa (japonica cultivar-group)}, partial (33%)
Length = 801
Score = 85.5 bits (210), Expect = 1e-17
Identities = 45/144 (31%), Positives = 83/144 (57%)
Frame = +1
Query: 64 ELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFRE 123
E+K F++FD + G I KELN ++ LG + ++++ QMI +D + G +D +EF
Sbjct: 136 EIKEAFELFDTDGSGTIDAKELNVAMRALGFEMTEEQIEQMIADVDKDGSGAIDYDEFEH 315
Query: 124 LYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIG 183
+ + + ERD +EE + +AF++ D++ +G IS +++ + LG Q T + ++M+
Sbjct: 316 MMTAKIGERDTKEE--LMKAFHIIDKDKNGKISASDIKRIAKELG--QNFTDREIQEMVD 483
Query: 184 TVDVDGNGLVDYKEFKQMMKGGGF 207
D + + VD +EF MM G+
Sbjct: 484 EADQNNDREVDPEEFIMMMNTTGY 555
Score = 57.8 bits (138), Expect = 3e-09
Identities = 27/69 (39%), Positives = 44/69 (63%)
Frame = +1
Query: 134 EEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLV 193
+++ ++++EAF +FD +G G I EL + +LG + T E ++MI VD DG+G +
Sbjct: 121 QQKRQEIKEAFELFDTDGSGTIDAKELNVAMRALGFEM--TEEQIEQMIADVDKDGSGAI 294
Query: 194 DYKEFKQMM 202
DY EF+ MM
Sbjct: 295 DYDEFEHMM 321
>TC78217 weakly similar to GP|20161870|dbj|BAB90783. hypothetical protein
{Oryza sativa (japonica cultivar-group)}, partial (49%)
Length = 1027
Score = 84.7 bits (208), Expect = 2e-17
Identities = 47/138 (34%), Positives = 80/138 (57%)
Frame = +2
Query: 66 KRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFRELY 125
+ V + FD + DG+++ EL L +G I KE IE +D + DG + +EE L
Sbjct: 230 EHVLRYFDEDGDGKVSPAELRQRLRIMGEEILLKEAEMAIEAMDSDGDGYLSLEELIALM 409
Query: 126 ESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTV 185
E +E++ +D+REAF ++D GFI+ L+ +L +G + +++++CK MI
Sbjct: 410 EE---GGEEQKLKDLREAFEMYDSEKCGFITPKSLKRMLKKMG--ESKSIDECKAMIKHF 574
Query: 186 DVDGNGLVDYKEFKQMMK 203
D+DG+GL+ + EF MM+
Sbjct: 575 DLDGDGLLSFDEFITMMQ 628
Score = 36.6 bits (83), Expect = 0.007
Identities = 21/73 (28%), Positives = 37/73 (49%)
Frame = +2
Query: 134 EEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLV 193
E + FD++GDG +S ELR L +G + +++ + I +D DG+G +
Sbjct: 209 EMKNAGFEHVLRYFDEDGDGKVSPAELRQRLRIMG--EEILLKEAEMAIEAMDSDGDGYL 382
Query: 194 DYKEFKQMMKGGG 206
+E +M+ GG
Sbjct: 383 SLEELIALMEEGG 421
>TC78218 weakly similar to GP|12963415|gb|AAK11255.1 regulator of gene
silencing {Nicotiana tabacum}, partial (58%)
Length = 793
Score = 84.3 bits (207), Expect = 3e-17
Identities = 47/138 (34%), Positives = 80/138 (57%)
Frame = +3
Query: 66 KRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFRELY 125
+ V + FD + DG+++ EL L +G I KE IE +D + DG + +EE L
Sbjct: 279 EHVLRYFDEDGDGKVSPAELRQRLRIMGEEILLKEAEMAIEAMDSDGDGYLSLEELIALM 458
Query: 126 ESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTV 185
E +E++ +D+REAF ++D GFI+ L+ +L +G + +++++CK MI
Sbjct: 459 EE---GGEEQKLKDLREAFEMYDSEKCGFITPKSLKRMLKKMG--ESKSIDECKAMIKHF 623
Query: 186 DVDGNGLVDYKEFKQMMK 203
D+DG+GL+ + EF MM+
Sbjct: 624 DLDGDGLLTFDEFITMMQ 677
>TC78949 similar to GP|16215471|emb|CAC82999. calcium-dependent protein
kinase 3 {Nicotiana tabacum}, partial (44%)
Length = 1015
Score = 81.6 bits (200), Expect = 2e-16
Identities = 45/141 (31%), Positives = 78/141 (54%)
Frame = +1
Query: 65 LKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFREL 124
LK +F+M D ++ G+IT +EL L+ +G + + E+ +++ DV+ G +D EF
Sbjct: 304 LKEMFKMIDTDNSGQITFEELKVGLKKVGANLKESEIYDLMQAADVDNSGTIDYGEF--- 474
Query: 125 YESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGT 184
+ + E E+ + AF+ FD++G G+I+ +EL+ G+K R E +I
Sbjct: 475 IAATLHINKIEREDHLFAAFSYFDKDGSGYITQEELQQACDEFGIKDVRLEE----IIKE 642
Query: 185 VDVDGNGLVDYKEFKQMMKGG 205
+D D +G +DY EF MM+ G
Sbjct: 643 IDEDNDGRIDYNEFAAMMQKG 705
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.139 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,265,231
Number of Sequences: 36976
Number of extensions: 65043
Number of successful extensions: 813
Number of sequences better than 10.0: 120
Number of HSP's better than 10.0 without gapping: 674
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 727
length of query: 211
length of database: 9,014,727
effective HSP length: 92
effective length of query: 119
effective length of database: 5,612,935
effective search space: 667939265
effective search space used: 667939265
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC146574.14


