

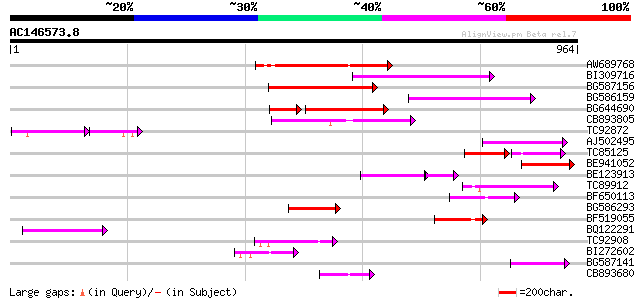
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146573.8 + phase: 0
(964 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 239 5e-63
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 181 1e-45
BG587156 similar to PIR|G85055|G8 probable polyprotein [imported... 176 5e-44
BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2... 173 3e-43
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 131 1e-39
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 145 1e-34
TC92872 92 2e-26
AJ502495 weakly similar to GP|18071369|g putative gag-pol polypr... 113 3e-25
TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-relate... 70 5e-25
BE941052 weakly similar to PIR|B85188|B85 retrotransposon like p... 111 1e-24
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 69 3e-20
TC89912 weakly similar to PIR|B84512|B84512 probable retroelemen... 97 3e-20
BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vu... 94 3e-19
BG586293 weakly similar to PIR|E84473|E84 probable retroelement ... 92 1e-18
BF519055 similar to GP|21592754|gb| unknown {Arabidopsis thalian... 91 2e-18
BQ122291 78 1e-14
TC92908 similar to GP|10140704|gb|AAG13538.1 putative gag-pol po... 78 2e-14
BI272602 GP|11602812|db avenaIII {Gallus gallus}, partial (3%) 71 2e-12
BG587141 similar to PIR|H86461|H86 hypothetical protein AAF32440... 66 6e-11
CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotia... 63 6e-10
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 239 bits (609), Expect = 5e-63
Identities = 115/233 (49%), Positives = 158/233 (67%)
Frame = +1
Query: 419 ITRSQNNIFKPKKFFYASNHPLPENLEPSNVRQAMQHPHWRQAISEEFNALIRNGTWSLV 478
ITR++ KPK F P P++ E + P W QA+ E+ ALI N TW LV
Sbjct: 1 ITRTKTGKLKPKVFL---TEPCPQDCE----NLPLSDPRWLQAMKTEYKALIDNKTWDLV 159
Query: 479 PPPKNKNIVDCKWLFRIKRNPDGTIDHYKARLVAKGFTQCPGVDFKETFAPVVRPQTIKL 538
P P +K + CKW++R+K NPDG+++ +KARLVAKGF+Q G D+ ETF+PV++P TI+L
Sbjct: 160 PLPPHKKAIGCKWVYRVKENPDGSVNKFKARLVAKGFSQTLGCDYTETFSPVIKPVTIRL 339
Query: 539 ILTIALTKGWKMHQLDVNNAFLQGSLNEEVYMAQPPGLKDSQQPTYVCKLHKAIYGLRQA 598
ILTIA+T W++ Q+D+NNAFL G L EEVYM+QP G ++ + VCKL+K++YGL+QA
Sbjct: 340 ILTIAITYKWEIQQIDINNAFLNGFLQEEVYMSQPQGF-EAANKSLVCKLNKSLYGLKQA 516
Query: 599 PRAWHDALKTFITSHGFTTSQSDPSLFIYASGTTLAYFLVYVDDLLLTGNDAS 651
PRAW++ L + GFT S+ DPSL IY Y +YVDD+L+TG+ AS
Sbjct: 517 PRAWYEXLTSAQIQFGFTKSRCDPSLLIYNQNGACIYLXIYVDDILITGSSAS 675
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 181 bits (459), Expect = 1e-45
Identities = 97/242 (40%), Positives = 139/242 (57%)
Frame = +2
Query: 583 TYVCKLHKAIYGLRQAPRAWHDALKTFITSHGFTTSQSDPSLFIYASGTTLAYFLVYVDD 642
T VC+L K+IYGL+QA R W+ L + S G+ S SD SLF ++ LVYVDD
Sbjct: 14 TKVCELQKSIYGLKQASRQWYSKLSESLISFGYLQSSSDFSLFTKFKDSSFTTLLVYVDD 193
Query: 643 LLLTGNDASFLHHFIQSLSNRFSLKHMGTPHYFLGIELIPSKTVLFLSQHKFIRDILQRF 702
++L GND S + H L +RF +K +G+ YFLG+E+ SK + L+Q K+ ++L+
Sbjct: 194 IVLAGNDISEIQHVKCFLIDRFKIKDLGSLRYFLGLEVARSKQGILLNQRKYTLELLEDS 373
Query: 703 DMDAAKPTYTPLSTTSTLTLNDGTAAADSTLYRQIIGALQYLNLTRPDLSFAINKLSQFM 762
A K T TP + L +D D T YR++IG L YL TRPD+SFA+ +LSQF+
Sbjct: 374 GNLAVKSTLTPYDISLKLHNSDSPLYNDETQYRRLIGKLIYLTTTRPDISFAVQQLSQFV 553
Query: 763 HKPTSLHFQHLKRLLRYLKATINFGILLKKTNALTLQVYTDADWDGNIDDRTSTSAYLIY 822
KP +H+Q R+L+YLK G+ T+ L L + D+DW R S + Y ++
Sbjct: 554 SKPQQVHYQAAIRVLQYLKTAPAKGLFYSATSNLKLSSFADSDWATCPTTRKSVTGYWVF 733
Query: 823 FG 824
G
Sbjct: 734 LG 739
>BG587156 similar to PIR|G85055|G8 probable polyprotein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 618
Score = 176 bits (445), Expect = 5e-44
Identities = 85/186 (45%), Positives = 120/186 (63%)
Frame = -1
Query: 440 LPENLEPSNVRQAMQHPHWRQAISEEFNALIRNGTWSLVPPPKNKNIVDCKWLFRIKRNP 499
L EN P + +AM+ W++++ E A+I+N TW PK K V +W+F IK
Sbjct: 576 LNENHIPRSYEEAMEDKEWKESVGAEAGAMIKNDTWYESELPKGKKAVSSRWIFTIKYKA 397
Query: 500 DGTIDHYKARLVAKGFTQCPGVDFKETFAPVVRPQTIKLILTIALTKGWKMHQLDVNNAF 559
DG+I+ K RLVA+GFT G D+ ETFAPV + TI+++L++A+ GW + Q+DV NAF
Sbjct: 396 DGSIERKKTRLVARGFTLTYGEDYIETFAPVAKLHTIRIVLSLAVNLGWGLWQMDVKNAF 217
Query: 560 LQGSLNEEVYMAQPPGLKDSQQPTYVCKLHKAIYGLRQAPRAWHDALKTFITSHGFTTSQ 619
LQG L +EVYM PPGL+ + V +L KAIYGL+Q+PRAW++ L T + GF S+
Sbjct: 216 LQGELEDEVYMYPPPGLEHLVKRGNVLRLKKAIYGLKQSPRAWYNKLSTTLNGRGFRKSE 37
Query: 620 SDPSLF 625
D +LF
Sbjct: 36 LDHTLF 19
>BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (11%)
Length = 732
Score = 173 bits (438), Expect = 3e-43
Identities = 82/217 (37%), Positives = 131/217 (59%)
Frame = +1
Query: 678 IELIPSKTVLFLSQHKFIRDILQRFDMDAAKPTYTPLSTTSTLTLNDGTAAADSTLYRQI 737
+E+I ++ +++ Q K++ D+L+RF M+ + + P++ L ++ D+T Y+QI
Sbjct: 1 VEVIQNEEGIYICQRKYVTDLLERFGMEKSNLSRNPIAPRCKLIKDENGVKVDATKYKQI 180
Query: 738 IGALQYLNLTRPDLSFAINKLSQFMHKPTSLHFQHLKRLLRYLKATINFGILLKKTNALT 797
+G L YL TRPDL + ++ +S+FM+ PT LH +KR+LRYL TIN GI+ K+ +
Sbjct: 181 VGCLMYLAATRPDLMYVLSLISRFMNCPTELHMHAVKRVLRYLNGTINLGIMYKRNGSEK 360
Query: 798 LQVYTDADWDGNIDDRTSTSAYLIYFGGNPISWLSRKQRTVARSSTEAEYRAVATATAEI 857
L+ YTD+D+ G++DDR STS Y+ +SW S+KQ V S+T+AE+ A A +
Sbjct: 361 LEAYTDSDYAGDLDDRKSTSGYVFMLSSGAVSWSSKKQPVVTLSTTKAEFIAAAFCACQS 540
Query: 858 MWITNLLSELHIPLSKPPLLLCDNVGATYLCSNPVLH 894
+W+ +L +L S + CDN L NPVLH
Sbjct: 541 VWMRRVLEKLGYTQSGSITMYCDNNSTIKLSKNPVLH 651
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 131 bits (330), Expect(2) = 1e-39
Identities = 58/141 (41%), Positives = 94/141 (66%)
Frame = -2
Query: 503 IDHYKARLVAKGFTQCPGVDFKETFAPVVRPQTIKLILTIALTKGWKMHQLDVNNAFLQG 562
I K++LV +G+ Q G+D+ E F+PV R + I++++ A G+K++Q+DV +AF+ G
Sbjct: 424 ITRNKSKLVVQGYNQKEGIDYDEAFSPVARMEAIRILIAFAAFMGFKLYQMDVKSAFING 245
Query: 563 SLNEEVYMAQPPGLKDSQQPTYVCKLHKAIYGLRQAPRAWHDALKTFITSHGFTTSQSDP 622
L EEV++ QPPG +D++ P +V +L+K +YGL+QAPRAW++ L F+ +GF + D
Sbjct: 244 DLKEEVFVKQPPGFEDAEVPNHVFRLNKTLYGLKQAPRAWYERLSKFLLKNGFKRGKIDN 65
Query: 623 SLFIYASGTTLAYFLVYVDDL 643
+LF+ L VYVDD+
Sbjct: 64 TLFLLKRE*ELLIIQVYVDDI 2
Score = 50.8 bits (120), Expect(2) = 1e-39
Identities = 19/54 (35%), Positives = 34/54 (62%)
Frame = -3
Query: 443 NLEPSNVRQAMQHPHWRQAISEEFNALIRNGTWSLVPPPKNKNIVDCKWLFRIK 496
++EP NV++A++ W ++ EE + R+ W LVP P+ K ++ +W+FR K
Sbjct: 594 SIEPKNVKEALRDADWINSMQEELHQFERSKVWYLVPRPEGKTVIGTRWVFRNK 433
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 145 bits (365), Expect = 1e-34
Identities = 80/255 (31%), Positives = 133/255 (51%), Gaps = 10/255 (3%)
Frame = +3
Query: 445 EPSNVRQAMQHPHWRQAISEEFNALIRNGTWSLVPPPKNKNIVDCKWLFRIKRNPDGTID 504
+P+ +A++ WR +++ E A RN TW L + KW+F+ K N +G I+
Sbjct: 39 DPTTFEEAVKSEKWRASMNNEMEATERNNTWELTDLRSGAKTIGLKWIFKTKLNENGEIE 218
Query: 505 HYKARLVAKGFTQCPGVDFKETFAPVVRPQTIKLILTIA---------LTKGWKMHQLDV 555
YKARLVAKG++Q GVD+ E FAPV R TI++++ +A ++ K H
Sbjct: 219 KYKARLVAKGYSQQYGVDYTEVFAPVARWDTIRMVIALAAQIKRDGVCIS*M*KAHSCME 398
Query: 556 NNAFLQGSLNEEVYMAQPPGLKDSQQPTYVCKLHKAIYGLRQAPRAWHDALKTFITSHGF 615
N +N V + ++ +A+YGL+QAPRAW+ ++ + T GF
Sbjct: 399 N*MRKFLLINHRVM*RR----------VIS*RVKRALYGLKQAPRAWYSRIEAYFTKEGF 548
Query: 616 TTSQSDPSLFIYAS-GTTLAYFLVYVDDLLLTGNDASFLHHFIQSLSNRFSLKHMGTPHY 674
+ +LF+ S G + +YVDDL+ GND + F +S+ F++ +G HY
Sbjct: 549 EKCPYEHTLFVKLSEGGKILIISLYVDDLIFIGNDENMFEEFKKSMKKEFNMSDLGKMHY 728
Query: 675 FLGIELIPSKTVLFL 689
FLG+E+ ++ +++
Sbjct: 729 FLGVEVTQNEKGIYI 773
>TC92872
Length = 923
Score = 92.0 bits (227), Expect(2) = 2e-26
Identities = 44/142 (30%), Positives = 81/142 (56%), Gaps = 9/142 (6%)
Frame = +1
Query: 3 QFNALLIGYDLFGFVDGTKPEPAEKHV---------DYNYWRRQDKLILHAILSSVEASI 53
Q LL G ++ G +DGT P E + DY W D+LI++ +LSS+ +
Sbjct: 142 QVTTLLAGIEVMGHIDGTTPIQTETIINNGVSAPNPDYTKWFTLDQLIINLLLSSMTEAD 321
Query: 54 ITMLGNVKNSKDAWDILNKMFASKTRARIMHLKERLTRTSKGSKSVSEYLQAIKAISDEL 113
+ + ++ W + F + +R+ +M + ++ R +KG KS+++YL ++K+++DEL
Sbjct: 322 SISFASYETARTLWVAIESQFNNTSRSHVMSVTNQIQRCTKGDKSITDYLFSVKSLADEL 501
Query: 114 AIINKPIDDDDLVIHAPNGLGS 135
A+I+K + DDD+ + NGLG+
Sbjct: 502 AVIDKSLSDDDITLFVLNGLGA 567
Score = 46.6 bits (109), Expect(2) = 2e-26
Identities = 32/118 (27%), Positives = 52/118 (43%), Gaps = 27/118 (22%)
Frame = +2
Query: 136 EFKEIAAALRTRENAIAFDELHDILVDHETFLQRD--QEPTIIPTAQVAYRGKPKYHKN- 192
E+ +IAA++RTR++ F+ELH L+ H+ +L+R+ Q +PTA A+ P +
Sbjct: 569 EYNDIAASIRTRQHPFTFEELHSHLLAHDDYLRREAVQVDIQVPTANFAHHTSPNQRSSK 748
Query: 193 ------------------GSFHNHINPSVNNIS------DSQGQQKQVMCQFCDKSGH 226
S H+ N +N +S+G + CQ C GH
Sbjct: 749 DDGILPIPSTGRGNNFSKQSQHHGSNTRGSNYRGRGRGFNSRGNRPPPRCQLCSLLGH 922
>AJ502495 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (9%)
Length = 542
Score = 113 bits (283), Expect = 3e-25
Identities = 51/144 (35%), Positives = 83/144 (57%)
Frame = +2
Query: 804 ADWDGNIDDRTSTSAYLIYFGGNPISWLSRKQRTVARSSTEAEYRAVATATAEIMWITNL 863
+DW G+ + R STS Y + G ISW S+KQ VA S+ EAEY A + + +W+ +
Sbjct: 2 SDWAGDTETRKSTSGYAFHLGTGAISWSSKKQPVVAFSTAEAEYIASTSCATQTVWLRRI 181
Query: 864 LSELHIPLSKPPLLLCDNVGATYLCSNPVLHSKMKHISLDYHFVREQVRDGKLQVSHVST 923
L +H + P + CDN A L NPV H + KHI + +H +RE + + ++ + + T
Sbjct: 182 LEVMHHEQNTPTKIYCDNKSAIALSKNPVFHGRSKHIDIQFHKIRELIAEKEVVIEYCPT 361
Query: 924 KDQLADLLTKPLPRAKFEELRSKM 947
++++AD+ TKPL F +L+ +
Sbjct: 362 EEKIADIFTKPLKIESFYKLKKML 433
>TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
(EC 3.4.23.-);, partial (7%)
Length = 705
Score = 69.7 bits (169), Expect(2) = 5e-25
Identities = 35/93 (37%), Positives = 51/93 (54%)
Frame = +3
Query: 853 ATAEIMWITNLLSELHIPLSKPPLLLCDNVGATYLCSNPVLHSKMKHISLDYHFVREQVR 912
A E +W+ L+ EL + + CD+ A ++ NP HS+ KHI + YHFVRE V
Sbjct: 240 ACKEAIWMQRLMEELGHK-QEQITVYCDSQSALHIARNPAFHSRTKHIGIQYHFVREVVE 416
Query: 913 DGKLQVSHVSTKDQLADLLTKPLPRAKFEELRS 945
+G + + + T D LAD +TK + KF RS
Sbjct: 417 EGSVDMQKIHTNDNLADAMTKSINTDKFIWCRS 515
Score = 63.9 bits (154), Expect(2) = 5e-25
Identities = 32/77 (41%), Positives = 48/77 (61%)
Frame = +1
Query: 773 LKRLLRYLKATINFGILLKKTNALTLQVYTDADWDGNIDDRTSTSAYLIYFGGNPISWLS 832
+KR++RY+K T + + LT++ Y D+D+ G+ D R ST+ Y+ G +SWLS
Sbjct: 1 VKRIMRYIKGTSGVAVCFGGSE-LTVRGYVDSDFAGDHDKRKSTTGYVFTLAGGAVSWLS 177
Query: 833 RKQRTVARSSTEAEYRA 849
+ Q VA S+TEAEY A
Sbjct: 178 KLQTVVALSTTEAEYMA 228
>BE941052 weakly similar to PIR|B85188|B85 retrotransposon like protein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 480
Score = 111 bits (278), Expect = 1e-24
Identities = 54/91 (59%), Positives = 69/91 (75%)
Frame = +2
Query: 870 PLSKPPLLLCDNVGATYLCSNPVLHSKMKHISLDYHFVREQVRDGKLQVSHVSTKDQLAD 929
P+S+ LL CD + ATYL NPV HS+MKHIS+D HFVR+ V+ GKL+V HV T DQLAD
Sbjct: 2 PISQSGLLRCDYLSATYLTHNPVYHSRMKHISIDIHFVRDLVQQGKLKVQHVCTVDQLAD 181
Query: 930 LLTKPLPRAKFEELRSKMKVTDGNFILRGHI 960
LTKPL +++ + LR+K+ VTDG ILRG +
Sbjct: 182 CLTKPLSKSRHQLLRNKIGVTDGTPILRGRV 274
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 68.9 bits (167), Expect(2) = 3e-20
Identities = 39/118 (33%), Positives = 59/118 (49%), Gaps = 1/118 (0%)
Frame = +1
Query: 597 QAPRAWHDALKTFITSHGFTTSQSDPSLFIYASGTTL-AYFLVYVDDLLLTGNDASFLHH 655
Q+PR W D + G+ Q+D ++FI S T A +VYVDD+ LTG+ +
Sbjct: 1 QSPRDWFDRFT*VVKKFGYIQCQTDHAMFIKHSSTVKKAILIVYVDDIFLTGDHGK*IKR 180
Query: 656 FIQSLSNRFSLKHMGTPHYFLGIELIPSKTVLFLSQHKFIRDILQRFDMDAAKPTYTP 713
L+ F +K +G YFLG+E+ K +SQ K++ D+L+ M K P
Sbjct: 181 LKNLLAEEFEIKDLGNLKYFLGMEVARWKKGSSISQRKYVLDLLKETRMIGCKTIRDP 354
Score = 48.9 bits (115), Expect(2) = 3e-20
Identities = 23/55 (41%), Positives = 33/55 (59%)
Frame = +2
Query: 708 KPTYTPLSTTSTLTLNDGTAAADSTLYRQIIGALQYLNLTRPDLSFAINKLSQFM 762
KP+ TP+ T L D D Y++++G L YL+ TRPD+SF + +SQFM
Sbjct: 338 KPSETPMDATVKLGTLDNGTLVDKGRYQRLVGKLIYLSHTRPDISFVVCTMSQFM 502
>TC89912 weakly similar to PIR|B84512|B84512 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(10%)
Length = 814
Score = 97.1 bits (240), Expect = 3e-20
Identities = 54/167 (32%), Positives = 91/167 (54%), Gaps = 4/167 (2%)
Frame = +1
Query: 771 QHLKRLLRYLKATINFGILLKKTNAL----TLQVYTDADWDGNIDDRTSTSAYLIYFGGN 826
Q LK +L+YL ++ LK T A L+ Y DAD+ GN+D R S S ++ G
Sbjct: 1 QALKWVLKYLNESLKSS--LKYTKAAQEEDALEGYVDADYAGNVDTRKSLSGFVFTLYGT 174
Query: 827 PISWLSRKQRTVARSSTEAEYRAVATATAEIMWITNLLSELHIPLSKPPLLLCDNVGATY 886
ISW + +Q V S+T+AEY A + +W+ ++ EL I + + CD+ A +
Sbjct: 175 TISWKANQQSVVTLSTTQAEYIAFVEGVKDAIWLKGMIGELGI-TQEYVKIHCDSQSAIH 351
Query: 887 LCSNPVLHSKMKHISLDYHFVREQVRDGKLQVSHVSTKDQLADLLTK 933
L ++ V H + KHI + HF+R+ + ++ V +++++ AD+ TK
Sbjct: 352 LANHQVYHERTKHIDIRLHFIRDMIESKEIVVEKMASEENPADVFTK 492
>BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vulgaris},
partial (13%)
Length = 494
Score = 93.6 bits (231), Expect = 3e-19
Identities = 48/123 (39%), Positives = 73/123 (59%), Gaps = 3/123 (2%)
Frame = +1
Query: 748 RPDLSFAINKLSQFMHKPTSLHFQHLKRLLRYLKATINFGILLK---KTNALTLQVYTDA 804
RPD+ ++++ +S+FMH P H R+LRY++ T+ +G+L K+ L Y+D+
Sbjct: 121 RPDICYSVSVISKFMHDPRKPHLIAANRILRYVRGTMEYGLLFPYGAKSEVYELICYSDS 300
Query: 805 DWDGNIDDRTSTSAYLIYFGGNPISWLSRKQRTVARSSTEAEYRAVATATAEIMWITNLL 864
DW G DR STS Y+ F ISW ++KQ A SS EAEY A AT + +W+ +++
Sbjct: 301 DWCG---DRRSTSGYVFKFNDAAISWCTKKQPITALSSYEAEYIAGTFATFQALWLDSVI 471
Query: 865 SEL 867
EL
Sbjct: 472 KEL 480
>BG586293 weakly similar to PIR|E84473|E84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 763
Score = 91.7 bits (226), Expect = 1e-18
Identities = 45/89 (50%), Positives = 59/89 (65%)
Frame = +2
Query: 474 TWSLVPPPKNKNIVDCKWLFRIKRNPDGTIDHYKARLVAKGFTQCPGVDFKETFAPVVRP 533
T LV P + +W+++IKRN DGT+ YKARLVAKG+ + G+DF E FAPVVR
Sbjct: 56 TLKLVKKPTGVKPIGLRWIYKIKRNEDGTLIKYKARLVAKGYVKQQGIDFDEVFAPVVRI 235
Query: 534 QTIKLILTIALTKGWKMHQLDVNNAFLQG 562
+TI L+L +A T G +H +DV AFL G
Sbjct: 236 ETI*LLLALAATNGC*IHHIDVKIAFLNG 322
>BF519055 similar to GP|21592754|gb| unknown {Arabidopsis thaliana}, partial
(30%)
Length = 675
Score = 91.3 bits (225), Expect = 2e-18
Identities = 47/91 (51%), Positives = 62/91 (67%)
Frame = -3
Query: 722 LNDGTAAADSTLYRQIIGALQYLNLTRPDLSFAINKLSQFMHKPTSLHFQHLKRLLRYLK 781
L DG+ ADS L I+GAL Y+ +T PDLSF+INK SQFMHKPT ++FQ LK+++R+ K
Sbjct: 670 LVDGSITADSKLVHSIVGALLYITVTCPDLSFSINKPSQFMHKPTQINFQQLKKVMRHPK 491
Query: 782 ATINFGILLKKTNALTLQVYTDADWDGNIDD 812
TI KK L + ++DADWD +I D
Sbjct: 490 LTI------KKLFDLQIYAFSDADWDDSIYD 416
>BQ122291
Length = 487
Score = 78.2 bits (191), Expect = 1e-14
Identities = 41/143 (28%), Positives = 74/143 (51%)
Frame = -2
Query: 23 EPAEKHVDYNYWRRQDKLILHAILSSVEASIITMLGNVKNSKDAWDILNKMFASKTRARI 82
E ++ Y W +D L+ ILS + S+++ +++S WD ++ ++ + R
Sbjct: 474 EAGTRNPAYTEWEDKDSLLCTWILSRISPSLLSRFVLLRHSWQVWDEIHSYCFTQMKTRS 295
Query: 83 MHLKERLTRTSKGSKSVSEYLQAIKAISDELAIINKPIDDDDLVIHAPNGLGSEFKEIAA 142
L+ L +KGS++VSE++ I+AIS+ LA I P+ DL+ L EF I A
Sbjct: 294 RQLRSELRSITKGSRTVSEFIARIRAISESLASIGDPVSHRDLIEVVLEALPEEFDPIVA 115
Query: 143 ALRTRENAIAFDELHDILVDHET 165
++ + ++ DEL L+ E+
Sbjct: 114 SVNAKSEVVSLDELESQLLTQES 46
>TC92908 similar to GP|10140704|gb|AAG13538.1 putative gag-pol polyprotein
{Oryza sativa}, partial (1%)
Length = 638
Score = 77.8 bits (190), Expect = 2e-14
Identities = 48/150 (32%), Positives = 79/150 (52%), Gaps = 9/150 (6%)
Frame = +2
Query: 416 NHIITRSQNN----IFKPKKFFYASNH-----PLPENLEPSNVRQAMQHPHWRQAISEEF 466
+H++T + ++ IF + +++ H + N+EP N QA Q W A+ +E
Sbjct: 164 SHLVTLNSSSKSYHIFHYVGYSFSAKHRASLAAITSNIEPKNYVQAAQ*QEWLAAMEQEI 343
Query: 467 NALIRNGTWSLVPPPKNKNIVDCKWLFRIKRNPDGTIDHYKARLVAKGFTQCPGVDFKET 526
L N T +L P + K VDC+ +++I +G I+ YKA+LVAK F Q G DF +
Sbjct: 344 QVLEENNTSTLEPLREGKKWVDCRPVYKIIHKANGEIEKYKAQLVAKDFVQVEGEDF*D- 520
Query: 527 FAPVVRPQTIKLILTIALTKGWKMHQLDVN 556
+ + +LTIA KG ++H +DV+
Sbjct: 521 LCLSNKDDNCRCLLTIAAAKG*QLHLMDVS 610
>BI272602 GP|11602812|db avenaIII {Gallus gallus}, partial (3%)
Length = 488
Score = 70.9 bits (172), Expect = 2e-12
Identities = 52/130 (40%), Positives = 70/130 (53%), Gaps = 22/130 (16%)
Frame = +1
Query: 383 SPSTSSGNE-------SSPSPSPTNNSIA--LTNP-----------PADSNATN--HIIT 420
SPS+ + NE SSP+ SPT+N +A T+P P +++ TN IIT
Sbjct: 22 SPSSPNSNEVAQNIQISSPA-SPTSNELANITTSPTPPPPPPPTARPHNNHTTNVGGIIT 198
Query: 421 RSQNNIFKPKKFFYASNHPLPENLEPSNVRQAMQHPHWRQAISEEFNALIRNGTWSLVPP 480
RSQNNI K K PL +EPSN+ QA++ WR + +EF+AL N TW LV
Sbjct: 199 RSQNNIVKTIKKLNLHVRPLCP-IEPSNITQALRDHDWRSIMQDEFDALQNNNTWDLVSR 375
Query: 481 PKNKNIVDCK 490
+N+V CK
Sbjct: 376 SFAQNLVGCK 405
Score = 32.3 bits (72), Expect = 0.90
Identities = 14/26 (53%), Positives = 18/26 (68%)
Frame = +2
Query: 477 LVPPPKNKNIVDCKWLFRIKRNPDGT 502
L+ P K +D W+FRIKRNPDG+
Sbjct: 365 LLAGPLLKIWLDVNWVFRIKRNPDGS 442
>BG587141 similar to PIR|H86461|H86 hypothetical protein AAF32440.1
[imported] - Arabidopsis thaliana, partial (20%)
Length = 731
Score = 66.2 bits (160), Expect = 6e-11
Identities = 36/100 (36%), Positives = 54/100 (54%)
Frame = +3
Query: 852 TATAEIMWITNLLSELHIPLSKPPLLLCDNVGATYLCSNPVLHSKMKHISLDYHFVREQV 911
TA + MW+ +LLSE+ + ++ DN L NPV H + HI YHF+RE V
Sbjct: 123 TAARQAMWLQDLLSEVTWEPCEEVVIRIDNQSVIALTRNPVFHGRGNHIHKRYHFIRECV 302
Query: 912 RDGKLQVSHVSTKDQLADLLTKPLPRAKFEELRSKMKVTD 951
+G+++V HV + A + TK L R F E+R + + D
Sbjct: 303 ENGQVEVEHVPGEKHRAYI*TKALGRIIFREIRYYIGMID 422
>CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotiana tabacum},
partial (7%)
Length = 780
Score = 62.8 bits (151), Expect = 6e-10
Identities = 37/94 (39%), Positives = 52/94 (54%)
Frame = -2
Query: 527 FAPVVRPQTIKLILTIALTKGWKMHQLDVNNAFLQGSLNEEVYMAQPPGLKDSQQPTYVC 586
F P+V+ TI +L+I + + LDV AFL+G L E++YM QP G + V
Sbjct: 554 FVPIVKLNTIMFLLSIVAIENLYLE*LDVKTAFLRGDLVEDIYMHQPEGF-S*EVGKMVG 378
Query: 587 KLHKAIYGLRQAPRAWHDALKTFITSHGFTTSQS 620
KL K++YGL+Q PR +LK T GF + S
Sbjct: 377 KLKKSMYGLKQGPRQCI*SLKALCTRKGFRSEIS 276
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 32,007,195
Number of Sequences: 36976
Number of extensions: 539109
Number of successful extensions: 5169
Number of sequences better than 10.0: 280
Number of HSP's better than 10.0 without gapping: 4059
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4808
length of query: 964
length of database: 9,014,727
effective HSP length: 105
effective length of query: 859
effective length of database: 5,132,247
effective search space: 4408600173
effective search space used: 4408600173
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146573.8


