

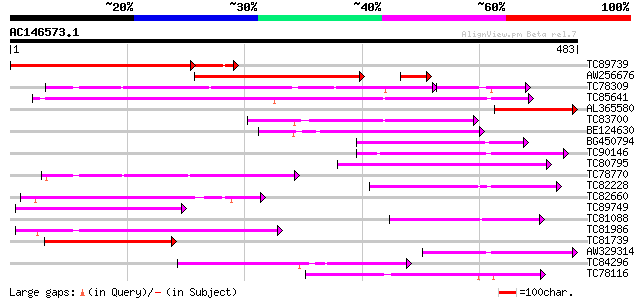
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146573.1 + phase: 0
(483 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC89739 similar to GP|9502380|gb|AAF88087.1| T12C24.27 {Arabidop... 330 1e-99
AW256676 similar to GP|9502380|gb| T12C24.27 {Arabidopsis thalia... 281 3e-76
TC78309 similar to GP|20259299|gb|AAM14385.1 putative cytochrome... 159 3e-54
TC85641 weakly similar to GP|8777295|dbj|BAA96885.1 cytochrome P... 202 2e-52
AL365580 similar to GP|6635860|gb|A cytochrome P450 {Helianthus ... 154 9e-38
TC83700 similar to GP|9587211|gb|AAF89209.1| cytochrome P450 {Vi... 142 4e-34
BE124630 weakly similar to GP|14209594|db cytochrome P450-like p... 114 1e-25
BG450794 similar to SP|O23051|C883 Cytochrome P450 88A3 (EC 1.14... 113 1e-25
TC90146 weakly similar to SP|Q42569|C901_ARATH Cytochrome P450 9... 110 1e-24
TC80795 weakly similar to GP|23495769|dbj|BAC19980. 5-alpha-taxa... 107 1e-23
TC78770 similar to GP|20259299|gb|AAM14385.1 putative cytochrome... 107 1e-23
TC82228 similar to GP|10177005|dbj|BAB10255. cytochrome P450 {Ar... 106 2e-23
TC82660 weakly similar to GP|22208463|gb|AAM94292.1 putative 5-a... 99 5e-21
TC89749 similar to PIR|T46143|T46143 steroid 22-alpha-hydroxylas... 98 8e-21
TC81088 weakly similar to GP|23495770|dbj|BAC19981. 5-alpha-taxa... 97 1e-20
TC81986 similar to GP|21537283|gb|AAM61624.1 cytochrome P450 pu... 93 3e-19
TC81739 similar to SP|Q42569|C901_ARATH Cytochrome P450 90A1 (EC... 93 3e-19
AW329314 weakly similar to SP|Q42569|C901_ Cytochrome P450 90A1 ... 92 4e-19
TC84296 similar to PIR|T46143|T46143 steroid 22-alpha-hydroxylas... 91 8e-19
TC78116 similar to PIR|F86441|F86441 probable cytochrome P450 [i... 87 1e-17
>TC89739 similar to GP|9502380|gb|AAF88087.1| T12C24.27 {Arabidopsis
thaliana}, partial (33%)
Length = 662
Score = 330 bits (845), Expect(2) = 1e-99
Identities = 155/158 (98%), Positives = 156/158 (98%)
Frame = +1
Query: 1 MWVLCLGALVTICIITRWVYRWRNPSCNGKLPPGSMGLPLLGESLQFFSPNTSCDIPPFI 60
MWVLCLGALVTICIITRWVYRWRNPSCNGKLPPGSMGLPLLGESLQFFSPNTSCDIPPFI
Sbjct: 82 MWVLCLGALVTICIITRWVYRWRNPSCNGKLPPGSMGLPLLGESLQFFSPNTSCDIPPFI 261
Query: 61 RKRMKRYGPIFKTNLVGRPVVVSTDPDLNYFIFQQEGKIFQSWYPDTFTEIFGQQNVGSL 120
RKRMKRYGPIFKTNLVGRPVVVSTDPDLNYFIFQQEGKIFQSWYPDTFTEIFGQQNVGSL
Sbjct: 262 RKRMKRYGPIFKTNLVGRPVVVSTDPDLNYFIFQQEGKIFQSWYPDTFTEIFGQQNVGSL 441
Query: 121 HGFMYKYLKNMMLNLFGPESLKKMISEVEQAACRTLQQ 158
HGFMYKYLKNMMLNLFGPESLKKMISEVEQAAC TL +
Sbjct: 442 HGFMYKYLKNMMLNLFGPESLKKMISEVEQAACXTLHK 555
Score = 52.0 bits (123), Expect(2) = 1e-99
Identities = 30/40 (75%), Positives = 31/40 (77%)
Frame = +3
Query: 156 LQQASCQDSVELKEATETMIFDLTAKKLISYDPTESSENL 195
+ QASC DSV LKEATETMIFDLTAK S TESSENL
Sbjct: 546 ITQASCLDSVXLKEATETMIFDLTAKN-SSL*STESSENL 662
>AW256676 similar to GP|9502380|gb| T12C24.27 {Arabidopsis thaliana}, partial
(29%)
Length = 639
Score = 281 bits (720), Expect = 3e-76
Identities = 144/145 (99%), Positives = 145/145 (99%)
Frame = -2
Query: 158 QASCQDSVELKEATETMIFDLTAKKLISYDPTESSENLRENFVAFIQGLISFPLNIPGTA 217
QASCQDSVELKEATETMIFDLTAK+LISYDPTESSENLRENFVAFIQGLISFPLNIPGTA
Sbjct: 638 QASCQDSVELKEATETMIFDLTAKQLISYDPTESSENLRENFVAFIQGLISFPLNIPGTA 459
Query: 218 YNKCLQGRKKAMKMLKNMLQERREMPRKQQMDFFDYVIEELRKEGTLLTEAIALDLMFVL 277
YNKCLQGRKKAMKMLKNMLQERREMPRKQQMDFFDYVIEELRKEGTLLTEAIALDLMFVL
Sbjct: 458 YNKCLQGRKKAMKMLKNMLQERREMPRKQQMDFFDYVIEELRKEGTLLTEAIALDLMFVL 279
Query: 278 LFASFETTSQALTYAIKLLSDNPLV 302
LFASFETTSQALTYAIKLLSDNPLV
Sbjct: 278 LFASFETTSQALTYAIKLLSDNPLV 204
Score = 49.7 bits (117), Expect = 3e-06
Identities = 24/26 (92%), Positives = 25/26 (95%)
Frame = +2
Query: 334 FTFQLITETARLANIVPGIFRKALRE 359
FTFQLITETARLANIVPGIFRKA R+
Sbjct: 137 FTFQLITETARLANIVPGIFRKAPRD 214
>TC78309 similar to GP|20259299|gb|AAM14385.1 putative cytochrome P450
protein {Arabidopsis thaliana}, partial (89%)
Length = 2063
Score = 159 bits (401), Expect(2) = 3e-54
Identities = 92/336 (27%), Positives = 187/336 (55%), Gaps = 2/336 (0%)
Frame = +1
Query: 31 LPPGSMGLPLLGESLQFFSPNTSCDIPPFIRKRMKRYGPIFKTNLVGRPVVVSTDPDLNY 90
LPPGSMG P +GE+ Q +S + + F ++KRYG +FK++++G P V+ + P+
Sbjct: 169 LPPGSMGWPYIGETFQLYSQDPNV----FFASKIKRYGSMFKSHILGCPCVMISSPEAAK 336
Query: 91 FIFQQEGKIFQSWYPDTFTEIFGQQNVGSLHGFMYKYLKNMMLNLFGPESLKKMISEVEQ 150
F+ + ++F+ +P + + G+Q + G + L+ ++L F P +++ ++ ++E
Sbjct: 337 FVLNK-AQLFKPTFPASKERMLGKQAIFFHQGEYHANLRRLVLRTFMPVAIRNIVPDIES 513
Query: 151 AACRTLQQASCQDSVELKEATETMIFDLTAKKLISYDPTESSENLRENFVAFIQGLISFP 210
A +L+ + E +T F++ + D E L++ + +G S P
Sbjct: 514 IAEDSLKSMEGRLITTFLEM-KTFTFNVALLSIFGKDEIHYREQLKQCYYTLEKGYNSMP 690
Query: 211 LNIPGTAYNKCLQGRKKAMKMLKNMLQERREMPRKQQMDFFDYVIEELRKEGTLLTEAIA 270
+N+PGT ++K ++ RK+ ++L ++ RRE ++ ++ D + + ++ L E IA
Sbjct: 691 INLPGTLFHKAMKARKELAQILAQIISSRRE----KKEEYKDLLGSFMDEKSGLSDEQIA 858
Query: 271 LDLMFVLLFASFETTSQALTYAIKLLSDNPLVFKQLQEEHEAILERRE--NPNSGVTWQE 328
+++ V +FA+ +TT+ LT+ +K L +N + + EE E+IL+ +E G+ W++
Sbjct: 859 NNVIGV-IFATRDTTASVLTWIVKYLGENISALESVIEEQESILKSKEENGEEKGLNWED 1035
Query: 329 YKSMTFTFQLITETARLANIVPGIFRKALREINFKG 364
K M T ++I ET R+A+I+ FR+A+ ++ ++G
Sbjct: 1036TKKMVITSRVIQETLRVASILSFTFREAVEDVEYQG 1143
Score = 71.6 bits (174), Expect(2) = 3e-54
Identities = 36/82 (43%), Positives = 49/82 (58%), Gaps = 2/82 (2%)
Frame = +3
Query: 364 GYTIPAGWAIMVCPPAVHLNPAKYQDPLVFNPSRWEGMEPSGATKH--FLAFGGGMRFCV 421
GY IP GW ++ +H +P ++DP F+PSR+E ATK F+ FG G+ C
Sbjct: 1251 GYLIPKGWKVLPLFRNIHHSPNNFKDPEKFDPSRFEA-----ATKPNTFMPFGSGIHACP 1415
Query: 422 GTEFAKVQMAVFLHCLVTKYRW 443
G E AK+++ V LH L TKYRW
Sbjct: 1416 GNELAKMEILVLLHHLTTKYRW 1481
>TC85641 weakly similar to GP|8777295|dbj|BAA96885.1 cytochrome P450-like
{Arabidopsis thaliana}, partial (71%)
Length = 1941
Score = 202 bits (515), Expect = 2e-52
Identities = 119/433 (27%), Positives = 218/433 (49%), Gaps = 6/433 (1%)
Frame = +1
Query: 20 YRWRNPSCNGKLPPGSMGLPLLGESLQFFSPNTSCDIPPFIRKRMKRYGP-IFKTNLVGR 78
Y+ ++P LPPG MG P++GESL+F S FI RM++Y +FKT++VG
Sbjct: 187 YKQKSPL---NLPPGKMGYPIIGESLEFLSTGWKGHPEKFIFDRMRKYSSELFKTSIVGE 357
Query: 79 PVVVSTDPDLNYFIFQQEGKIFQSWYPDTFTEIFGQQNVGSLHGFMYKYLKNMMLNLFGP 138
VV N F+F E K+ +W+PD+ +IF ++ S ++ ++ F P
Sbjct: 358 STVVCCGAASNKFLFSNENKLVTAWWPDSVNKIFPTTSLDSNLKEESIKMRKLLPQFFKP 537
Query: 139 ESLKKMISEVEQAACRT-LQQASCQDSVELKEATETMIFDLTAKKLISYDPTESSENLRE 197
E+L++ + ++ A R + ++ + + + F L + +S + +
Sbjct: 538 EALQRYVGVMDVIAQRHFVTHWDNKNEITVYPLAKRYTFLLACRLFMSVEDENHVAKFSD 717
Query: 198 NFVAFIQGLISFPLNIPGTAYNKCLQG----RKKAMKMLKNMLQERREMPRKQQMDFFDY 253
F G+IS P+++PGT +NK ++ RK+ +K++K + E D +
Sbjct: 718 PFQLIAAGIISLPIDLPGTPFNKAIKASNFIRKELIKIIKQRRVDLAEGTASPTQDILSH 897
Query: 254 VIEELRKEGTLLTEAIALDLMFVLLFASFETTSQALTYAIKLLSDNPLVFKQLQEEHEAI 313
++ + G + E D + LL +T S A T+ +K L + P ++ ++ +E I
Sbjct: 898 MLLTSDENGKSMNELNIADKILGLLIGGHDTASVACTFLVKYLGELPHIYDKVYQEQMEI 1077
Query: 314 LERRENPNSGVTWQEYKSMTFTFQLITETARLANIVPGIFRKALREINFKGYTIPAGWAI 373
+ + + W + K M +++ + E RL+ + G FR+A+ + F G++IP GW +
Sbjct: 1078AKSKP-AGELLNWDDLKKMKYSWNVACEVMRLSPPLQGGFREAITDFMFNGFSIPKGWKL 1254
Query: 374 MVCPPAVHLNPAKYQDPLVFNPSRWEGMEPSGATKHFLAFGGGMRFCVGTEFAKVQMAVF 433
+ H N + P F+P+R+EG P+ T F+ FGGG R C G E+A++++ VF
Sbjct: 1255YWSANSTHKNAECFPMPEKFDPTRFEGNGPAPYT--FVPFGGGPRMCPGKEYARLEILVF 1428
Query: 434 LHCLVTKYRWRPI 446
+H LV +++W +
Sbjct: 1429MHNLVKRFKWEKV 1467
>AL365580 similar to GP|6635860|gb|A cytochrome P450 {Helianthus annuus},
partial (25%)
Length = 516
Score = 154 bits (388), Expect = 9e-38
Identities = 70/70 (100%), Positives = 70/70 (100%)
Frame = +1
Query: 414 GGGMRFCVGTEFAKVQMAVFLHCLVTKYRWRPIKGGNIVRTPGLQFPNGFHVQITEKDQK 473
GGGMRFCVGTEFAKVQMAVFLHCLVTKYRWRPIKGGNIVRTPGLQFPNGFHVQITEKDQK
Sbjct: 1 GGGMRFCVGTEFAKVQMAVFLHCLVTKYRWRPIKGGNIVRTPGLQFPNGFHVQITEKDQK 180
Query: 474 KHESECTTTY 483
KHESECTTTY
Sbjct: 181 KHESECTTTY 210
>TC83700 similar to GP|9587211|gb|AAF89209.1| cytochrome P450 {Vigna
radiata}, partial (42%)
Length = 658
Score = 142 bits (357), Expect = 4e-34
Identities = 77/202 (38%), Positives = 114/202 (56%), Gaps = 5/202 (2%)
Frame = +3
Query: 203 IQGLISFPLNIPGTAYNKCLQGRKKAMKMLKNMLQERRE-----MPRKQQMDFFDYVIEE 257
I+G + P + T Y + ++ R K + L ++++RR+ + +K M +
Sbjct: 3 IEGFFTLPFPLFSTTYRRAIKARTKVAEALTLVVRQRRKENEIGIQKKNDM------LGA 164
Query: 258 LRKEGTLLTEAIALDLMFVLLFASFETTSQALTYAIKLLSDNPLVFKQLQEEHEAILERR 317
L G + +D M LL A +ETTS +T AIK L++ PL QL+EEHE I R
Sbjct: 165 LLASGEQFSNEEIVDFMLALLVAGYETTSTIMTLAIKFLTETPLALAQLKEEHEQIRARS 344
Query: 318 ENPNSGVTWQEYKSMTFTFQLITETARLANIVPGIFRKALREINFKGYTIPAGWAIMVCP 377
E P + + W +YKSMTFT ++ ET R+ANI+ IFR+ +I+ KGYTIP G ++
Sbjct: 345 E-PEAALEWTDYKSMTFTQCVVNETLRVANIIGAIFRRTTTDIDIKGYTIPKGMKVIASF 521
Query: 378 PAVHLNPAKYQDPLVFNPSRWE 399
AVHLNP ++D FNP RW+
Sbjct: 522 RAVHLNPEHFKDARTFNPWRWQ 587
>BE124630 weakly similar to GP|14209594|db cytochrome P450-like protein
{Oryza sativa (japonica cultivar-group)}, partial (33%)
Length = 608
Score = 114 bits (284), Expect = 1e-25
Identities = 63/197 (31%), Positives = 108/197 (53%), Gaps = 5/197 (2%)
Frame = +1
Query: 213 IPGTAYNKCLQGRKKAMKMLKNMLQERR-----EMPRKQQMDFFDYVIEELRKEGTLLTE 267
+PGT + LQ +KK +K ++ +Q +R E+PR D D ++ + ++ LT+
Sbjct: 1 LPGTKLYQSLQAKKKMVK*VRKTVQAKRNKGIFEVPR----DVVDVLLNDTSEK---LTD 159
Query: 268 AIALDLMFVLLFASFETTSQALTYAIKLLSDNPLVFKQLQEEHEAILERRENPNSGVTWQ 327
+ D + ++ ++ +T A K LS+ P +QL E+ + + ++ + W
Sbjct: 160 DLIADNIIDMMIPGEDSVPVLMTLATKYLSECPPALQQLTVENIKLKKLKDQLGKPLCWN 339
Query: 328 EYKSMTFTFQLITETARLANIVPGIFRKALREINFKGYTIPAGWAIMVCPPAVHLNPAKY 387
+Y S+ FT ++ITET R+ NI+ G+ RKAL+++ KGY IP GW + +VHL+ Y
Sbjct: 340 DYLSLPFTQKVITETLRMGNIINGVMRKALKDVEIKGYLIPQGWCVFANFRSVHLDEKNY 519
Query: 388 QDPLVFNPSRWEGMEPS 404
+ P FNP RW+ E S
Sbjct: 520 ECPYQFNPWRWQVREKS 570
>BG450794 similar to SP|O23051|C883 Cytochrome P450 88A3 (EC 1.14.-.-).
[Mouse-ear cress] {Arabidopsis thaliana}, partial (33%)
Length = 691
Score = 113 bits (283), Expect = 1e-25
Identities = 55/147 (37%), Positives = 85/147 (57%)
Frame = +1
Query: 296 LSDNPLVFKQLQEEHEAILERRENPNSGVTWQEYKSMTFTFQLITETARLANIVPGIFRK 355
L +P ++ +EE E I++RR G+T +E + M F +++I ET R+ +FR+
Sbjct: 4 LQKHPEYLQKAKEEQEEIIKRRPPTQKGLTLKEIRGMDFLYKVIDETMRVITFSLVVFRE 183
Query: 356 ALREINFKGYTIPAGWAIMVCPPAVHLNPAKYQDPLVFNPSRWEGMEPSGATKHFLAFGG 415
A ++ GYTIP GW ++ +VHL+P Y +P FNP+RW +G FL FG
Sbjct: 184 AKSDVMINGYTIPKGWKVLTWFRSVHLDPEIYPNPKEFNPNRWNKEHKAG---EFLPFGA 354
Query: 416 GMRFCVGTEFAKVQMAVFLHCLVTKYR 442
G R C G + AK+++AVFLH + Y+
Sbjct: 355 GTRLCPGNDLAKMEIAVFLHHFILNYQ 435
>TC90146 weakly similar to SP|Q42569|C901_ARATH Cytochrome P450 90A1 (EC
1.14.-.-). [Mouse-ear cress] {Arabidopsis thaliana},
partial (7%)
Length = 777
Score = 110 bits (275), Expect = 1e-24
Identities = 59/182 (32%), Positives = 100/182 (54%), Gaps = 1/182 (0%)
Frame = +2
Query: 296 LSDNPLVFKQLQEEHEAILERRENPNSGVTWQEYKSMTFTFQLITETARLANIVPGIFRK 355
L+ P+V K+L+EE+ A R+ +P +T ++ ++ +T +++ E R+ANI FRK
Sbjct: 2 LAKYPIVLKKLREENMAF--RKGSPEDFITPKDVSNLKYTNKVVEEVIRMANIAACAFRK 175
Query: 356 ALREINFKGYTIPAGWAIMVCPPAVHLNPAKYQDPLVFNPSRWEGMEPSGATKHFLAFGG 415
E+++KGY IP GW +++ +H + ++DP+ FNP RW G + FGG
Sbjct: 176 VDTEVDYKGYKIPKGWNVILLLRYLHTDSKNFKDPMNFNPDRWNEPAKPGT---YQPFGG 346
Query: 416 GMRFCVGTEFAKVQMAVFLHCLVTKYRWRPIK-GGNIVRTPGLQFPNGFHVQITEKDQKK 474
G R C G A++Q+A+ LH L Y+W I +I+ +G V+ ++ +K
Sbjct: 347 GQRLCPGNTLARIQLALLLHHLSIGYKWELINPNADIIYLSHPAPVDGVEVKFSKL*RKN 526
Query: 475 HE 476
HE
Sbjct: 527 HE 532
>TC80795 weakly similar to GP|23495769|dbj|BAC19980.
5-alpha-taxadienol-10-beta-hydroxylase-like protein
{Oryza sativa (japonica cultivar-group)}, partial (12%)
Length = 796
Score = 107 bits (267), Expect = 1e-23
Identities = 55/182 (30%), Positives = 94/182 (51%)
Frame = +3
Query: 280 ASFETTSQALTYAIKLLSDNPLVFKQLQEEHEAILERRENPNSGVTWQEYKSMTFTFQLI 339
AS +T++ ++ I LS + V+ ++ EE IL++RE G+TW E + M +T+++
Sbjct: 6 ASHDTSATLMSLMIWKLSRDQEVYNKVLEEQMEILKQREANEEGLTWGEIQKMKYTWRVA 185
Query: 340 TETARLANIVPGIFRKALREINFKGYTIPAGWAIMVCPPAVHLNPAKYQDPLVFNPSRWE 399
E R+ + G FRKAL++ ++GY IP GW + H N +++P F PSR+E
Sbjct: 186 QELMRMIPPLFGAFRKALKDTCYQGYDIPKGWQVYWASSGTHENKDIFENPYKFEPSRFE 365
Query: 400 GMEPSGATKHFLAFGGGMRFCVGTEFAKVQMAVFLHCLVTKYRWRPIKGGNIVRTPGLQF 459
+L FG G+ C+G EFA+V+ +H V W + + + +
Sbjct: 366 NQTKPIPPFTYLPFGAGLHNCIGNEFARVETLTTIHNFVKMCEWSQLNPEETITRQPMPY 545
Query: 460 PN 461
P+
Sbjct: 546 PS 551
>TC78770 similar to GP|20259299|gb|AAM14385.1 putative cytochrome P450
protein {Arabidopsis thaliana}, partial (43%)
Length = 1007
Score = 107 bits (267), Expect = 1e-23
Identities = 62/223 (27%), Positives = 121/223 (53%), Gaps = 3/223 (1%)
Frame = +1
Query: 28 NGK---LPPGSMGLPLLGESLQFFSPNTSCDIPPFIRKRMKRYGPIFKTNLVGRPVVVST 84
NGK LPPGSMG P +GE+ Q +S + S F ++KRYG +FK++++G P V+ +
Sbjct: 310 NGKQLPLPPGSMGYPYIGETFQMYSQDPSL----FFANKIKRYGAMFKSHILGCPCVMIS 477
Query: 85 DPDLNYFIFQQEGKIFQSWYPDTFTEIFGQQNVGSLHGFMYKYLKNMMLNLFGPESLKKM 144
P+ F+ + ++F+ +P + + G+Q + G + L+ ++L F PE++K +
Sbjct: 478 SPEAAKFVLNKS-QLFKPTFPASKERMLGKQAIFFHQGNYHANLRRLVLRSFMPEAIKSI 654
Query: 145 ISEVEQAACRTLQQASCQDSVELKEATETMIFDLTAKKLISYDPTESSENLRENFVAFIQ 204
+ +E A +T ++ + + +T F++ + D E+L+ + +
Sbjct: 655 VPNIESIA-QTCLKSWDGNLITTYLEMKTFTFNVALLSIFGKDEILYREDLKRCYYTLEK 831
Query: 205 GLISFPLNIPGTAYNKCLQGRKKAMKMLKNMLQERREMPRKQQ 247
G S P+N+PGT ++K ++ RK+ ++L+ ++ RR +K Q
Sbjct: 832 GYNSMPINLPGTLFHKAMKARKELAQILEQIISTRRCKKQKLQ 960
>TC82228 similar to GP|10177005|dbj|BAB10255. cytochrome P450 {Arabidopsis
thaliana}, partial (30%)
Length = 1003
Score = 106 bits (265), Expect = 2e-23
Identities = 57/166 (34%), Positives = 95/166 (56%), Gaps = 2/166 (1%)
Frame = +3
Query: 307 QEEHEAILERRENPNS-GVTWQEYKSMTFTFQLITETARLANIVPGIFRKALREINFKGY 365
QE+ I ++EN G+ W++ K+M T ++I ET R+A+I+ FR+A ++ ++GY
Sbjct: 6 QEQMSIIKGKQENGEEIGLNWEDTKNMPITSRVIQETLRVASILSFTFREATEDVEYQGY 185
Query: 366 TIPAGWAIMVCPPAVHLNPAKYQDPLVFNPSRWEGMEPSGATKHFLAFGGGMRFCVGTEF 425
IP GW ++ +H +P +++P F+PSR+E + P T F+ FG G+ C G E
Sbjct: 186 LIPKGWKVLPLFRNIHHSPENFKEPEKFDPSRFE-VAPKPNT--FMPFGNGVHACPGNEL 356
Query: 426 AKVQMAVFLHCLVTKYRWRPIKGGNIVRTPGLQFP-NGFHVQITEK 470
AK+++ V +H L TKYRW + N ++ P NG + + K
Sbjct: 357 AKLEILVLVHHLTTKYRWSVVGEKNGIQYGPFALPQNGLPINLYSK 494
>TC82660 weakly similar to GP|22208463|gb|AAM94292.1 putative
5-alpha-taxadienol-10-beta-hydroxylase {Sorghum
bicolor}, partial (15%)
Length = 780
Score = 98.6 bits (244), Expect = 5e-21
Identities = 63/222 (28%), Positives = 105/222 (46%), Gaps = 13/222 (5%)
Frame = +3
Query: 10 VTICIITRWVY----RWRNPSCNGKLPPGSMGLPLLGESLQFFSPNTSCDIPPFIRKRMK 65
+ IC++T + + + S +P GS+G P++GE+L F +I +R+
Sbjct: 105 IAICVLTLSLVFLLKKILSKSQTKNVPKGSLGYPIIGETLNFLKAQRQVKGSEWIEERVS 284
Query: 66 RYGPIFKTNLVGRPVVVSTDPDLNYFIFQQEGKIFQSWYPDTFTEIFGQQNVGSLHGFMY 125
+YGP+FKT+L+G P V N FI + + P T +I G+Q + L G +
Sbjct: 285 KYGPVFKTSLLGSPTVFIIGQQGNKFILGSSDDVLSANKPPTIQKILGKQTLAELIGSRH 464
Query: 126 KYLKNMMLNLFGPESLKKMISEVEQAACRTLQQASCQDSVELKEATETMIFDLTAKKLIS 185
+ LK ML PE L+ + ++++ + L + ELKE T + L K +S
Sbjct: 465 RLLKGEMLKFLKPECLQNYVKKMDELVYKVLLK-------ELKENKTTQVVRLMKK--LS 617
Query: 186 YD---------PTESSENLRENFVAFIQGLISFPLNIPGTAY 218
YD + E L E+F + + S P+N PGT++
Sbjct: 618 YDMACNVLFDIDDHTREILFEDFTTAFKAIHSLPINFPGTSF 743
>TC89749 similar to PIR|T46143|T46143 steroid 22-alpha-hydroxylase (DWF4) -
Arabidopsis thaliana, partial (29%)
Length = 661
Score = 97.8 bits (242), Expect = 8e-21
Identities = 50/146 (34%), Positives = 86/146 (58%), Gaps = 1/146 (0%)
Frame = +1
Query: 6 LGALVTICIITRWVYRWRNPSCNGKLPPGSMGLPLLGESLQFFSPNTSCDIPPFIRKRMK 65
L +++++ I ++ + + N LPPG MG P +GE++ + P ++ I F+ + +
Sbjct: 142 LSSILSLLIFIFFLIKTKQAKPNLNLPPGRMGWPFIGETIGYLKPYSATTIGKFMEQHIA 321
Query: 66 RYGPIFKTNLVGRPVVVSTDPDLNYFIFQQEGKIFQSWYPDTFTEIFGQQNVGSLHGFMY 125
RYG I+K+NL G P +VS D LN FI Q EGK+F+ YP + I G+ ++ L G M+
Sbjct: 322 RYGKIYKSNLFGGPTIVSADAGLNRFILQNEGKLFECSYPSSIGGILGKWSMLVLVGDMH 501
Query: 126 KYLKNMMLNLFGPESLK-KMISEVEQ 150
+ ++N+ LN L+ ++ EVE+
Sbjct: 502 RDMRNISLNFLCHARLRTHLLKEVEK 579
>TC81088 weakly similar to GP|23495770|dbj|BAC19981.
5-alpha-taxadienol-10-beta-hydroxylase-like protein
{Oryza sativa (japonica cultivar-group)}, partial (28%)
Length = 724
Score = 97.4 bits (241), Expect = 1e-20
Identities = 49/132 (37%), Positives = 74/132 (55%)
Frame = +3
Query: 324 VTWQEYKSMTFTFQLITETARLANIVPGIFRKALREINFKGYTIPAGWAIMVCPPAVHLN 383
+TW++ M FT+++ ET R + + G FRK +I + GY IP GW I H++
Sbjct: 45 LTWEDLSKMKFTWRVAMETLRRFSPIFGGFRKTATDIEYGGYIIPKGWQIFWVTSMTHMD 224
Query: 384 PAKYQDPLVFNPSRWEGMEPSGATKHFLAFGGGMRFCVGTEFAKVQMAVFLHCLVTKYRW 443
+ +P F+PSR+E + S F+ FGGG R C G EFA+V+ V +H LVTK+ W
Sbjct: 225 NNIFPEPSKFDPSRFENL-ASTPPYCFVPFGGGARICPGYEFARVETLVAIHYLVTKFSW 401
Query: 444 RPIKGGNIVRTP 455
+ + + R P
Sbjct: 402 KLLSENSFSRDP 437
>TC81986 similar to GP|21537283|gb|AAM61624.1 cytochrome P450 putative
{Arabidopsis thaliana}, partial (21%)
Length = 797
Score = 92.8 bits (229), Expect = 3e-19
Identities = 59/233 (25%), Positives = 110/233 (46%), Gaps = 6/233 (2%)
Frame = +3
Query: 6 LGALVTICIITRWVYRW-----RNPSCNGKLPPGSMGLPLLGESLQFFSPNTSCDIPPFI 60
L ++ + ++T + +++ + + KLPPGSMG P +G++LQ +S + + F
Sbjct: 111 LAYIIVLFLLTLFSFKFFPKRPKTKQTSAKLPPGSMGWPYIGQTLQLYSQDPNV----FF 278
Query: 61 RKRMKRYGPIFKTNLVGRPVVVSTDPDLNYFIFQQEGKIFQSWYPDTFTEIFGQQNVGSL 120
+ KRYG IFKTN++G V+ P+ F+ + +F+ YP + + G +
Sbjct: 279 FSKQKRYGEIFKTNILGCKCVMLASPEAARFVLVTQSHLFKPTYPKSKERLIGPCALFFH 458
Query: 121 HGFMYKYLKNMMLNLFGPESLKKMISEVEQAACRTLQQASCQD-SVELKEATETMIFDLT 179
G + L+ ++ +SL+ ++ E+E A T++ + + + F++
Sbjct: 459 QGEYHLRLRKLIQRSLSLDSLRNLVPEIEALAVSTIKSWGDDGCMINTFKEMKKFSFEVG 638
Query: 180 AKKLISYDPTESSENLRENFVAFIQGLISFPLNIPGTAYNKCLQGRKKAMKML 232
K+ E L++N+ G SF IPGT Y K L R+K +L
Sbjct: 639 ILKVFGNLEPRLREELKKNYWIVDNGYNSFSTQIPGTQYKKALLAREKLGSIL 797
>TC81739 similar to SP|Q42569|C901_ARATH Cytochrome P450 90A1 (EC 1.14.-.-).
[Mouse-ear cress] {Arabidopsis thaliana}, partial (35%)
Length = 656
Score = 92.8 bits (229), Expect = 3e-19
Identities = 42/113 (37%), Positives = 70/113 (61%)
Frame = +1
Query: 30 KLPPGSMGLPLLGESLQFFSPNTSCDIPPFIRKRMKRYGPIFKTNLVGRPVVVSTDPDLN 89
+LPPGS+GLP +GE+LQ S S + PFI +R+ RYG IF +++ G P V S DP+ N
Sbjct: 190 QLPPGSLGLPFIGETLQMISAYKSDNPEPFIDQRVNRYGSIFTSHVFGEPTVFSADPETN 369
Query: 90 YFIFQQEGKIFQSWYPDTFTEIFGQQNVGSLHGFMYKYLKNMMLNLFGPESLK 142
FI EGK+F+ YP + + + G+ ++ + G ++K + ++ ++ +K
Sbjct: 370 RFIIMNEGKLFECSYPGSISNLLGKHSLLLMKGSLHKKMHSLTMSFANSSIIK 528
>AW329314 weakly similar to SP|Q42569|C901_ Cytochrome P450 90A1 (EC
1.14.-.-). [Mouse-ear cress] {Arabidopsis thaliana},
partial (9%)
Length = 651
Score = 92.0 bits (227), Expect = 4e-19
Identities = 47/133 (35%), Positives = 71/133 (53%), Gaps = 1/133 (0%)
Frame = +1
Query: 352 IFRKALREINFKGYTIPAGWAIMVCPPAVHLNPAKYQDPLVFNPSRWEGMEPSGATKHFL 411
+FRKA E+++KGY IP GW ++V +H +P ++DP+ FNP RW SG +
Sbjct: 4 VFRKAANEVDYKGYKIPKGWNVIVLLRYLHTDPENFKDPMYFNPDRWNEPAMSGT---YQ 174
Query: 412 AFGGGMRFCVGTEFAKVQMAVFLHCLVTKYRWRPIK-GGNIVRTPGLQFPNGFHVQITEK 470
FG G R C G A++Q+A+ LH L Y+W I +IV +G V+ +
Sbjct: 175 VFGAGQRLCPGNMLARIQLALLLHHLSIGYKWELINPNADIVYLSHPAPVDGVEVKFSRL 354
Query: 471 DQKKHESECTTTY 483
+K HE+ +Y
Sbjct: 355 *RKNHENWIVFSY 393
>TC84296 similar to PIR|T46143|T46143 steroid 22-alpha-hydroxylase (DWF4) -
Arabidopsis thaliana, partial (36%)
Length = 679
Score = 91.3 bits (225), Expect = 8e-19
Identities = 61/206 (29%), Positives = 109/206 (52%), Gaps = 7/206 (3%)
Frame = +1
Query: 144 MISEVEQAACRTLQQASCQDSVELKEATETMIFDLTAKKLISYDPTE-SSENLRENFVAF 202
++ EVE+ L + + ++ + F+L A+ ++S P + +ENL++ +V F
Sbjct: 76 LLKEVEKHTRLVLSSWKEKTTFAAQDEAKKFTFNLMAEHIMSLQPGKIETENLKKEYVTF 255
Query: 203 IQGLISFPLNIPGTAYNKCLQGRKKAMKMLKNMLQERREMPRK-----QQMDFFDYVIEE 257
++G++S PLN PGTAY + L+ R +K ++ ++ER + ++ ++ D ++V+
Sbjct: 256 MKGVVSAPLNFPGTAYWRALKSRCTILKFIEGKMEERMKRMQEGNENSEENDLLNWVL-- 429
Query: 258 LRKEGTLLTEAIALDLMFVLLFASFETTSQALTYAIKLLSDNPLVFKQLQEEHEAILERR 317
K L TE I LDL+ LLFA ET+S ++ AI L P QL+EEH+ I +
Sbjct: 430 --KHSNLSTEQI-LDLILSLLFAGHETSSVSIALAIYFLPGCPQAILQLREEHKEIARAK 600
Query: 318 ENPNSGVT-WQEYKSMTFTFQLITET 342
+ T + M FT ++ +T
Sbjct: 601 KTSRGNRTDLGRLQKMEFTHCVVNKT 678
>TC78116 similar to PIR|F86441|F86441 probable cytochrome P450 [imported] -
Arabidopsis thaliana, partial (80%)
Length = 1547
Score = 87.0 bits (214), Expect = 1e-17
Identities = 63/209 (30%), Positives = 100/209 (47%), Gaps = 5/209 (2%)
Frame = +3
Query: 253 YVIEELRKEGTLLTEAIALDLMFVLLFASFETTSQALTYAIKLLSDNPLVFKQLQEEHEA 312
Y L G +T D + +L A ET++ LT+ LLS P V +LQEE ++
Sbjct: 879 YFFTFLLASGDDVTSKQLRDDLMTMLIAGHETSAAVLTWTFYLLSKEPSVMSKLQEEVDS 1058
Query: 313 ILERRENPNSGVTWQEYKSMTFTFQLITETARLANIVPGIFRKALREINFKGYTIPAGWA 372
+L R T ++ K + +T ++I E+ RL P + R+++ + Y I G
Sbjct: 1059 VLGDRFP-----TIEDMKKLKYTTRVINESLRLYPQPPVLIRRSIEDDVLGEYPIKRGED 1223
Query: 373 IMVCPPAVHLNPAKYQDPLVFNPSRW--EGMEPSGATKHF--LAFGGGMRFCVGTEFAKV 428
I + +H +P + D F P RW +G P+ + F L FGGG R C+G FA
Sbjct: 1224 IFISVWNLHRSPTLWNDADKFEPERWPLDGPNPNETNQGFKYLPFGGGPRKCIGDMFASY 1403
Query: 429 QMAVFLHCLVTKYRWRPIKGG-NIVRTPG 456
++ V L LV ++ ++ G +V T G
Sbjct: 1404 EVVVALAMLVRRFNFQMAVGAPPVVMTTG 1490
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.138 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,566,201
Number of Sequences: 36976
Number of extensions: 222613
Number of successful extensions: 1258
Number of sequences better than 10.0: 167
Number of HSP's better than 10.0 without gapping: 1198
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1209
length of query: 483
length of database: 9,014,727
effective HSP length: 100
effective length of query: 383
effective length of database: 5,317,127
effective search space: 2036459641
effective search space used: 2036459641
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146573.1


