

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146571.1 - phase: 0
(713 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
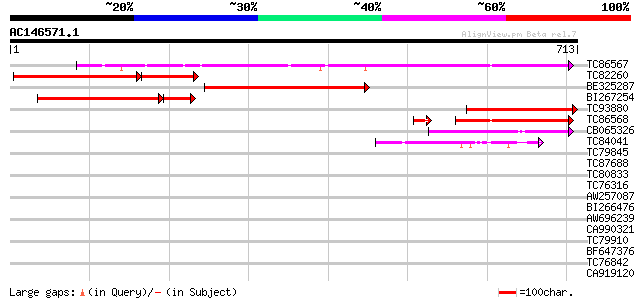
Score E
Sequences producing significant alignments: (bits) Value
TC86567 similar to GP|15384989|emb|CAC59823. Xaa-Pro aminopeptid... 480 e-136
TC82260 similar to GP|19310478|gb|AAL84973.1 AT3g05350/T12H1_32 ... 318 e-124
BE325287 weakly similar to GP|19310478|gb AT3g05350/T12H1_32 {Ar... 409 e-114
BI267254 similar to GP|19310478|gb AT3g05350/T12H1_32 {Arabidops... 322 e-106
TC93880 similar to GP|19310478|gb|AAL84973.1 AT3g05350/T12H1_32 ... 293 1e-79
TC86568 similar to GP|15384991|emb|CAC59824. Xaa-Pro aminopeptid... 158 2e-42
CB065326 similar to PIR|D81858|D81 probable aminopeptidase NMA16... 141 1e-33
TC84041 similar to PIR|A86226|A86226 hypothetical protein [impor... 42 8e-04
TC79845 similar to GP|21554069|gb|AAM63150.1 ethylene responsive... 36 0.059
TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~un... 35 0.100
TC80833 weakly similar to PIR|D86175|D86175 hypothetical protein... 32 0.65
TC76316 weakly similar to GP|11994454|dbj|BAB02456. contains sim... 32 1.1
AW257087 homologue to GP|6683126|dbj KIAA0339 protein {Homo sapi... 31 1.4
BI266476 weakly similar to PIR|T50661|T50 receptor-type protein ... 31 1.9
AW696239 homologue to PIR|D71441|D714 hypothetical protein - Ara... 30 2.5
CA990321 weakly similar to PIR|S03170|S03 homeotic protein cut -... 30 3.2
TC79910 similar to GP|9293979|dbj|BAB01882.1 gene_id:MAL21.25~un... 30 4.2
BF647376 similar to GP|7939533|dbj| Nicotiana EREBP-3-like prote... 30 4.2
TC76842 similar to GP|8809606|dbj|BAA97157.1 ethylene responsive... 30 4.2
CA919120 PIR|F84614|F8 probable kinesin heavy chain [imported] -... 30 4.2
>TC86567 similar to GP|15384989|emb|CAC59823. Xaa-Pro aminopeptidase 1
{Lycopersicon esculentum}, partial (91%)
Length = 2223
Score = 480 bits (1235), Expect = e-136
Identities = 276/663 (41%), Positives = 383/663 (57%), Gaps = 39/663 (5%)
Frame = +1
Query: 85 LTALRRLFSKPDVSIDAYIIPSQDAHQSEFIAQSYARRKYISAFTGSNGTAVVTN----D 140
L+ALR L S + A ++PS+D HQSE+++ AR K + +V +
Sbjct: 55 LSALRSLMSSHSPPLHALVVPSEDYHQSEYVS---ARDKQSCICFWIHMEVLVWHL*QRM 225
Query: 141 KAALWTDGRYFLQAEKQLNSNWILMRAGNPGVPTTSEWLNEVLAPGARVGIDPFLFTSDA 200
K LWTDGRYFLQAE+QL+ W LMR P W+ + L A +G+DP+ + D
Sbjct: 226 KHLLWTDGRYFLQAEQQLSDQWKLMRLAED--PAVDIWMADNLPKDAAIGVDPWCISIDT 399
Query: 201 AEELKHVISKKNHELVYLYNSNLVDEIWKEARPEPPNKPVRVHGLKYAGLDVASKLSSLR 260
A+ + +KK +LV NLVDE+W P N V V LK+AG V KL LR
Sbjct: 400 AQRWERAFAKKQQKLVQT-TKNLVDEVWTTRPPAEINAAV-VQPLKFAGRSVTDKLKDLR 573
Query: 261 SELVQAGSSAIIVTALDEIAWLLNLRGSDIPHSPVVYAYLIVEIDGAKLFIDNSKVTEEV 320
+L Q + I++TALDE+AWL N+RG D+ + PVV+A+ IV + A +++D KV+ EV
Sbjct: 574 KKLAQEHARGIVLTALDEVAWLYNIRGKDVAYCPVVHAFAIVTSNSAFIYVDKRKVSIEV 753
Query: 321 DDHLKKANIEIRPYNSIVSEIENLAARGSSLWLDTSSVNAAIVNAYKAACD-RYYQNYES 379
HL++ IEI+ Y + + LA S+ A++ K + + ++
Sbjct: 754 KTHLEENGIEIKEYTEVSLDAAFLATNELD---SVSTAKASLAEVTKQSENSETNKSVNG 924
Query: 380 KHKTRSKGFD-------------GSIANSDVPIAVHKSSPVSLAKAIKNETELKGMQECH 426
KH+T K + S N D V + SP++L KA+KN EL G+++ H
Sbjct: 925 KHQTGEKCSNLIWADPASCCYALYSKLNPDA--VVLQQSPLALPKALKNPVELDGLRKAH 1098
Query: 427 LRDAAALAQFWDWLEKEITN---------------------DRILTEVEVSDKLLEFRSK 465
+RD AA+ Q+ WL+ ++ + LTEV VSDKL EFR+
Sbjct: 1099VRDGAAVVQYLVWLDNKMQDIYGASGYFLEENTVKKEKPLKSLKLTEVTVSDKLEEFRAS 1278
Query: 466 QAGFLDTSFDTISGSGPNGAIIHYKPEPGSCSTVDANKLFLLDSGAQYVDGTTDITRTVH 525
+ F SF TIS GPN AIIHY P+ +C+ +D +K++L DSGAQY+DGTTDITRTVH
Sbjct: 1279KEHFRGLSFPTISSVGPNAAIIHYSPQAETCAELDPDKIYLFDSGAQYLDGTTDITRTVH 1458
Query: 526 FGKPTTREKECFTRVLQGHIALDQAVFPEDTPGFVLDAFARSFLWKVGLDYRHGTGHGVG 585
FG+P+ EK C+T VL+GHIAL A FP T G LD +R LW GLDYRHGTGHG+G
Sbjct: 1459FGRPSDHEKACYTAVLKGHIALGNARFPNGTNGHQLDILSRIPLWNYGLDYRHGTGHGIG 1638
Query: 586 AALNVHEGPQGISYRYGNLTPLVNGMIVSNEPGYYEDHAFGIRIENLLYVRNVETPNRFG 645
+ LNVHEGP IS+R N+ PL M V++EPGYYED AFGIR+EN+L + +T FG
Sbjct: 1639SYLNVHEGPHLISFRIRNV-PLQASMTVTDEPGYYEDGAFGIRLENVLVINEADTKFNFG 1815
Query: 646 GIQYLGFEKLTYVPIQIKLVDVSLLSTTEIDWLNNYHSVVWEKVSPLLDGSARQWLWNNT 705
YL FE +T+ P Q KL+D++LL+ E +WLN+YHS + ++P LD + WL T
Sbjct: 1816DKGYLSFEHITWAPYQTKLIDLNLLNPEEKNWLNSYHSKCRDILAPHLDEAGNAWLKKAT 1995
Query: 706 QPI 708
+P+
Sbjct: 1996EPV 2004
>TC82260 similar to GP|19310478|gb|AAL84973.1 AT3g05350/T12H1_32
{Arabidopsis thaliana}, partial (20%)
Length = 700
Score = 318 bits (816), Expect(2) = e-124
Identities = 160/160 (100%), Positives = 160/160 (100%)
Frame = +2
Query: 6 LSSSSSSSPATVTKLALSIHSSFPTLNLNPIFFYKFKSNKHNKPPFFSIRNCTTSNSISA 65
LSSSSSSSPATVTKLALSIHSSFPTLNLNPIFFYKFKSNKHNKPPFFSIRNCTTSNSISA
Sbjct: 2 LSSSSSSSPATVTKLALSIHSSFPTLNLNPIFFYKFKSNKHNKPPFFSIRNCTTSNSISA 181
Query: 66 KPSSQLRKNRSSNSDSDPKLTALRRLFSKPDVSIDAYIIPSQDAHQSEFIAQSYARRKYI 125
KPSSQLRKNRSSNSDSDPKLTALRRLFSKPDVSIDAYIIPSQDAHQSEFIAQSYARRKYI
Sbjct: 182 KPSSQLRKNRSSNSDSDPKLTALRRLFSKPDVSIDAYIIPSQDAHQSEFIAQSYARRKYI 361
Query: 126 SAFTGSNGTAVVTNDKAALWTDGRYFLQAEKQLNSNWILM 165
SAFTGSNGTAVVTNDKAALWTDGRYFLQAEKQLNSNWILM
Sbjct: 362 SAFTGSNGTAVVTNDKAALWTDGRYFLQAEKQLNSNWILM 481
Score = 146 bits (369), Expect(2) = e-124
Identities = 70/72 (97%), Positives = 70/72 (97%)
Frame = +3
Query: 166 RAGNPGVPTTSEWLNEVLAPGARVGIDPFLFTSDAAEELKHVISKKNHELVYLYNSNLVD 225
RAGNPGVPTTSEWLNEVLAPGARVGIDPFLFTSDAAEELKHVISKKNHELVYLYNSNLVD
Sbjct: 483 RAGNPGVPTTSEWLNEVLAPGARVGIDPFLFTSDAAEELKHVISKKNHELVYLYNSNLVD 662
Query: 226 EIWKEARPEPPN 237
EIWKEAR EP N
Sbjct: 663 EIWKEARAEPSN 698
>BE325287 weakly similar to GP|19310478|gb AT3g05350/T12H1_32 {Arabidopsis
thaliana}, partial (24%)
Length = 630
Score = 409 bits (1051), Expect = e-114
Identities = 208/208 (100%), Positives = 208/208 (100%)
Frame = +1
Query: 245 LKYAGLDVASKLSSLRSELVQAGSSAIIVTALDEIAWLLNLRGSDIPHSPVVYAYLIVEI 304
LKYAGLDVASKLSSLRSELVQAGSSAIIVTALDEIAWLLNLRGSDIPHSPVVYAYLIVEI
Sbjct: 1 LKYAGLDVASKLSSLRSELVQAGSSAIIVTALDEIAWLLNLRGSDIPHSPVVYAYLIVEI 180
Query: 305 DGAKLFIDNSKVTEEVDDHLKKANIEIRPYNSIVSEIENLAARGSSLWLDTSSVNAAIVN 364
DGAKLFIDNSKVTEEVDDHLKKANIEIRPYNSIVSEIENLAARGSSLWLDTSSVNAAIVN
Sbjct: 181 DGAKLFIDNSKVTEEVDDHLKKANIEIRPYNSIVSEIENLAARGSSLWLDTSSVNAAIVN 360
Query: 365 AYKAACDRYYQNYESKHKTRSKGFDGSIANSDVPIAVHKSSPVSLAKAIKNETELKGMQE 424
AYKAACDRYYQNYESKHKTRSKGFDGSIANSDVPIAVHKSSPVSLAKAIKNETELKGMQE
Sbjct: 361 AYKAACDRYYQNYESKHKTRSKGFDGSIANSDVPIAVHKSSPVSLAKAIKNETELKGMQE 540
Query: 425 CHLRDAAALAQFWDWLEKEITNDRILTE 452
CHLRDAAALAQFWDWLEKEITNDRILTE
Sbjct: 541 CHLRDAAALAQFWDWLEKEITNDRILTE 624
>BI267254 similar to GP|19310478|gb AT3g05350/T12H1_32 {Arabidopsis
thaliana}, partial (21%)
Length = 620
Score = 322 bits (824), Expect(2) = e-106
Identities = 158/158 (100%), Positives = 158/158 (100%)
Frame = +1
Query: 36 IFFYKFKSNKHNKPPFFSIRNCTTSNSISAKPSSQLRKNRSSNSDSDPKLTALRRLFSKP 95
IFFYKFKSNKHNKPPFFSIRNCTTSNSISAKPSSQLRKNRSSNSDSDPKLTALRRLFSKP
Sbjct: 1 IFFYKFKSNKHNKPPFFSIRNCTTSNSISAKPSSQLRKNRSSNSDSDPKLTALRRLFSKP 180
Query: 96 DVSIDAYIIPSQDAHQSEFIAQSYARRKYISAFTGSNGTAVVTNDKAALWTDGRYFLQAE 155
DVSIDAYIIPSQDAHQSEFIAQSYARRKYISAFTGSNGTAVVTNDKAALWTDGRYFLQAE
Sbjct: 181 DVSIDAYIIPSQDAHQSEFIAQSYARRKYISAFTGSNGTAVVTNDKAALWTDGRYFLQAE 360
Query: 156 KQLNSNWILMRAGNPGVPTTSEWLNEVLAPGARVGIDP 193
KQLNSNWILMRAGNPGVPTTSEWLNEVLAPGARVGIDP
Sbjct: 361 KQLNSNWILMRAGNPGVPTTSEWLNEVLAPGARVGIDP 474
Score = 82.8 bits (203), Expect(2) = e-106
Identities = 39/40 (97%), Positives = 39/40 (97%)
Frame = +2
Query: 194 FLFTSDAAEELKHVISKKNHELVYLYNSNLVDEIWKEARP 233
FLFTSDAAE LKHVISKKNHELVYLYNSNLVDEIWKEARP
Sbjct: 482 FLFTSDAAEXLKHVISKKNHELVYLYNSNLVDEIWKEARP 601
>TC93880 similar to GP|19310478|gb|AAL84973.1 AT3g05350/T12H1_32
{Arabidopsis thaliana}, partial (18%)
Length = 728
Score = 293 bits (751), Expect = 1e-79
Identities = 139/139 (100%), Positives = 139/139 (100%)
Frame = +1
Query: 575 DYRHGTGHGVGAALNVHEGPQGISYRYGNLTPLVNGMIVSNEPGYYEDHAFGIRIENLLY 634
DYRHGTGHGVGAALNVHEGPQGISYRYGNLTPLVNGMIVSNEPGYYEDHAFGIRIENLLY
Sbjct: 1 DYRHGTGHGVGAALNVHEGPQGISYRYGNLTPLVNGMIVSNEPGYYEDHAFGIRIENLLY 180
Query: 635 VRNVETPNRFGGIQYLGFEKLTYVPIQIKLVDVSLLSTTEIDWLNNYHSVVWEKVSPLLD 694
VRNVETPNRFGGIQYLGFEKLTYVPIQIKLVDVSLLSTTEIDWLNNYHSVVWEKVSPLLD
Sbjct: 181 VRNVETPNRFGGIQYLGFEKLTYVPIQIKLVDVSLLSTTEIDWLNNYHSVVWEKVSPLLD 360
Query: 695 GSARQWLWNNTQPIIRETV 713
GSARQWLWNNTQPIIRETV
Sbjct: 361 GSARQWLWNNTQPIIRETV 417
>TC86568 similar to GP|15384991|emb|CAC59824. Xaa-Pro aminopeptidase 2
{Lycopersicon esculentum}, partial (27%)
Length = 823
Score = 158 bits (399), Expect(2) = 2e-42
Identities = 76/148 (51%), Positives = 101/148 (67%)
Frame = +3
Query: 561 LDAFARSFLWKVGLDYRHGTGHGVGAALNVHEGPQGISYRYGNLTPLVNGMIVSNEPGYY 620
LD +R LW GLDYRHGTGHG+G+ LNVHEGP IS+R N+ PL M V++EPGYY
Sbjct: 120 LDILSRIPLWNYGLDYRHGTGHGIGSYLNVHEGPHLISFRIRNV-PLQASMTVTDEPGYY 296
Query: 621 EDHAFGIRIENLLYVRNVETPNRFGGIQYLGFEKLTYVPIQIKLVDVSLLSTTEIDWLNN 680
ED AFGIR+EN+L + +T FG YL FE +T+ P Q KL+D++LL+ E +WLN+
Sbjct: 297 EDGAFGIRLENVLVINEADTKFNFGDKGYLSFEHITWAPYQTKLIDLNLLNPEEKNWLNS 476
Query: 681 YHSVVWEKVSPLLDGSARQWLWNNTQPI 708
YHS + ++P LD + WL T+P+
Sbjct: 477 YHSKCRDILAPHLDEAGNAWLKKATEPV 560
Score = 33.5 bits (75), Expect(2) = 2e-42
Identities = 17/22 (77%), Positives = 18/22 (81%)
Frame = +2
Query: 509 SGAQYVDGTTDITRTVHFGKPT 530
SGAQY+DGTTDITRT G PT
Sbjct: 62 SGAQYLDGTTDITRT--NGPPT 121
>CB065326 similar to PIR|D81858|D81 probable aminopeptidase NMA1640
[imported] - Neisseria meningitidis (strain Z2491
serogroup A), partial (19%)
Length = 577
Score = 141 bits (355), Expect = 1e-33
Identities = 83/184 (45%), Positives = 107/184 (58%), Gaps = 2/184 (1%)
Frame = +2
Query: 527 GKPTTREKECFTRVLQGHIALDQAVFPEDTPGFVLDAFARSFLWKVGLDYRHGTGHGVGA 586
G+P+ +K TRVL+G L A FP +LDA AR+ +W +DY HGTGHGVG
Sbjct: 5 GQPSLEQKRDCTRVLKGLNGLASATFPRGILSPLLDAIARAPIWADLVDYGHGTGHGVGY 184
Query: 587 ALNVHEGPQGISYRYGNL--TPLVNGMIVSNEPGYYEDHAFGIRIENLLYVRNVETPNRF 644
LNVHEGPQ I+Y+ T + GMI S EPG Y +G+RIENL+ R + + F
Sbjct: 185 FLNVHEGPQVIAYQAATTPQTAMQVGMISSIEPGTYRPGEWGVRIENLVVNREAGS-SVF 361
Query: 645 GGIQYLGFEKLTYVPIQIKLVDVSLLSTTEIDWLNNYHSVVWEKVSPLLDGSARQWLWNN 704
G +L FE LT PI + + LL+ E DWLN YH V E+++PLL G A WL
Sbjct: 362 G--DFLNFETLTLCPIDTRCLQPELLAKEERDWLNEYHRRVRERLAPLLSGDALAWLEAR 535
Query: 705 TQPI 708
TQ +
Sbjct: 536 TQAL 547
>TC84041 similar to PIR|A86226|A86226 hypothetical protein [imported] -
Arabidopsis thaliana, partial (54%)
Length = 901
Score = 42.0 bits (97), Expect = 8e-04
Identities = 56/237 (23%), Positives = 99/237 (41%), Gaps = 25/237 (10%)
Frame = +1
Query: 460 LEFRSKQAGFLDTSFDTISGSGPNGAIIHYKPEPGSCSTVDANKLFLLDSGAQYVDGTTD 519
+E+ K G F+ + G GPNG++IHY + L L+D G + +D
Sbjct: 226 VEYECKMRGAQRMGFNPVVGGGPNGSVIHYSRND---QKIQDGDLVLMDIGCELHGYLSD 396
Query: 520 ITRT-VHFGKPTTREKECFTRVLQGHIALDQAVFPEDTPGFVLDAFAR---------SFL 569
+TRT G ++ ++E + +L+ + + P + + + L
Sbjct: 397 LTRTWPPCGSFSSAQEELYELILETNKHCVELCKPGASIRQIHNHSVEMLQKGLKEVGIL 576
Query: 570 WKVGLDYRH-----GTGHGVGAALNVHEGPQGISYRYGNLTPLVNGMIVSNEPGYYEDHA 624
VG H GH +G +++H+ IS+ PL G++++ EPG Y +
Sbjct: 577 KDVGSSSYHKLNPTSIGHYLG--MDIHD-CSAISFD----CPLKPGVVITIEPGVYIPSS 735
Query: 625 F---------GIRIENLLYVRNVETPNRFGGIQYLGFEKLT-YVPIQIKLVDVSLLS 671
F GIRIE+ + + G+E LT +P ++K + SLL+
Sbjct: 736 FNCPERYQGIGIRIEDEILITET------------GYEVLTASIPKEVKQIQ-SLLN 867
>TC79845 similar to GP|21554069|gb|AAM63150.1 ethylene responsive element
binding factor-like {Arabidopsis thaliana}, partial
(39%)
Length = 857
Score = 35.8 bits (81), Expect = 0.059
Identities = 29/122 (23%), Positives = 51/122 (41%), Gaps = 3/122 (2%)
Frame = +1
Query: 303 EIDGAKLFIDNSKVTEE---VDDHLKKANIEIRPYNSIVSEIENLAARGSSLWLDTSSVN 359
E+ A + + KV +E VD+ + + RP+ +EI + + +GS +WL T
Sbjct: 286 EVAIATTMVSSKKVKKEEECVDETRRYRGVRRRPWGKFAAEIRDPSRKGSRVWLGTFDCE 465
Query: 360 AAIVNAYKAACDRYYQNYESKHKTRSKGFDGSIANSDVPIAVHKSSPVSLAKAIKNETEL 419
AY +A R +G +I N + V P S + + E+E+
Sbjct: 466 IDAAKAYDSAA------------FRMRG-QKAILNFPLEAGVCDPKPNSCGRKRRRESEI 606
Query: 420 KG 421
+G
Sbjct: 607 EG 612
>TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~unknown
protein {Arabidopsis thaliana}, partial (46%)
Length = 2077
Score = 35.0 bits (79), Expect = 0.100
Identities = 39/135 (28%), Positives = 61/135 (44%), Gaps = 11/135 (8%)
Frame = -2
Query: 5 SLSSSSSSSPATVTKLALSIHSSFPTLNLNPI-----FFYKFK------SNKHNKPPFFS 53
S SSS S+SPA + + S+ SS +L+ +P F K+ N + PP +
Sbjct: 729 SSSSSISNSPAFSSSSSSSLSSSASSLS-SPFS*LSSSFSKYSVNSSIFKNLSSIPPPLT 553
Query: 54 IRNCTTSNSISAKPSSQLRKNRSSNSDSDPKLTALRRLFSKPDVSIDAYIIPSQDAHQSE 113
++S+S S+ PSS + SS+ S P L FS P S + S + +S
Sbjct: 552 FSFSSSSSSSSSLPSSS--SSSSSSKSSSPSLPPFAFFFSTPSKSSSS---SSNSSSKSS 388
Query: 114 FIAQSYARRKYISAF 128
+ S + R + S F
Sbjct: 387 LSSSSNSTRFFFSNF 343
>TC80833 weakly similar to PIR|D86175|D86175 hypothetical protein [imported]
- Arabidopsis thaliana, partial (60%)
Length = 709
Score = 32.3 bits (72), Expect = 0.65
Identities = 15/48 (31%), Positives = 22/48 (45%)
Frame = +1
Query: 322 DHLKKANIEIRPYNSIVSEIENLAARGSSLWLDTSSVNAAIVNAYKAA 369
D +K I RP+ +EI + +G+ +WL T AY AA
Sbjct: 166 DQIKYRGIRRRPWGKFAAEIRDPTRKGTRIWLGTFDTAEQAARAYDAA 309
>TC76316 weakly similar to GP|11994454|dbj|BAB02456. contains similarity to
receptor protein kinase~gene_id:MLD14.4 {Arabidopsis
thaliana}, partial (37%)
Length = 1745
Score = 31.6 bits (70), Expect = 1.1
Identities = 21/55 (38%), Positives = 32/55 (58%), Gaps = 7/55 (12%)
Frame = +3
Query: 436 FWDWLEKEITNDRILTEVEVSDKLL--EFR-----SKQAGFLDTSFDTISGSGPN 483
F+ + KEITN + E+++S+ L EF SKQ FLD F+ ++GS P+
Sbjct: 531 FYGSIPKEITNYKYFYELDLSNNKLVGEFPKEVLDSKQLVFLDLRFNQLTGSVPS 695
>AW257087 homologue to GP|6683126|dbj KIAA0339 protein {Homo sapiens},
partial (2%)
Length = 466
Score = 31.2 bits (69), Expect = 1.4
Identities = 25/78 (32%), Positives = 40/78 (51%)
Frame = +1
Query: 5 SLSSSSSSSPATVTKLALSIHSSFPTLNLNPIFFYKFKSNKHNKPPFFSIRNCTTSNSIS 64
S SSS+SSS ++V K A+S + P K +S+ + S + ++S+S S
Sbjct: 187 SSSSSTSSSVSSVKKPAVSAAKATPA--------KKEESDSDSDSDSSSSSSSSSSSSSS 342
Query: 65 AKPSSQLRKNRSSNSDSD 82
+ SS + SSNSD +
Sbjct: 343 SSSSSSSSSSSSSNSDDE 396
>BI266476 weakly similar to PIR|T50661|T50 receptor-type protein kinase LRK1
[imported] - Arabidopsis thaliana, partial (27%)
Length = 661
Score = 30.8 bits (68), Expect = 1.9
Identities = 25/121 (20%), Positives = 52/121 (42%), Gaps = 3/121 (2%)
Frame = +1
Query: 513 YVDGTTDITRT--VHFGKPTTREKECFTRVLQGHIALDQAVFPEDTPGFVLDAFARSFLW 570
Y+DG ++T + T R+K GH + ++T + +F+ +FL+
Sbjct: 55 YLDGIAELTTNGLLKLTNDTERDK--------GHGFYPNPIVFKNTSSESVSSFSTTFLF 210
Query: 571 KVGLDYRHGTGHGVGAALNVHEG-PQGISYRYGNLTPLVNGMIVSNEPGYYEDHAFGIRI 629
+ Y + GHG+ ++ +G P + +Y G+ + G +H FG+ +
Sbjct: 211 AIIPQYANIGGHGIVFVISPTKGLPDSLPSQY-------LGLFNDSNSGNSSNHVFGVEL 369
Query: 630 E 630
+
Sbjct: 370 D 372
>AW696239 homologue to PIR|D71441|D714 hypothetical protein - Arabidopsis
thaliana, partial (9%)
Length = 621
Score = 30.4 bits (67), Expect = 2.5
Identities = 27/95 (28%), Positives = 44/95 (45%), Gaps = 1/95 (1%)
Frame = +3
Query: 1 MNMRSLSSSSSSSPATVTKLALSIHSSFPTLNLNPIFFYKFKSNKHNKPPFFSIRNCTTS 60
MN++ SSSSSS + K S + PT ++P K+ NKP S+ TT+
Sbjct: 120 MNLKHPKSSSSSSSRAIVKAKPS--NVAPTTKVDPTLTKMVKNFMQNKPK--SVNTTTTT 287
Query: 61 NSISAKPSSQLRKNRSSNSDSDPKLTAL-RRLFSK 94
+ PS + K+ ++ + L ++LF K
Sbjct: 288 TKLFI-PSDVIAKDLKKDAKRVTGFSTLQKKLFGK 389
>CA990321 weakly similar to PIR|S03170|S03 homeotic protein cut - fruit fly
(Drosophila melanogaster), partial (2%)
Length = 451
Score = 30.0 bits (66), Expect = 3.2
Identities = 22/57 (38%), Positives = 27/57 (46%), Gaps = 4/57 (7%)
Frame = -1
Query: 5 SLSSSSSS----SPATVTKLALSIHSSFPTLNLNPIFFYKFKSNKHNKPPFFSIRNC 57
S +SSSSS S T+T I S LNLN +F K+ K + FS NC
Sbjct: 382 SFTSSSSSILLFSSTTLTFFFSHIDDSSRILNLNTVFSICVKTRKSSSSKEFSSLNC 212
>TC79910 similar to GP|9293979|dbj|BAB01882.1 gene_id:MAL21.25~unknown
protein {Arabidopsis thaliana}, partial (67%)
Length = 741
Score = 29.6 bits (65), Expect = 4.2
Identities = 27/92 (29%), Positives = 36/92 (38%)
Frame = +3
Query: 3 MRSLSSSSSSSPATVTKLALSIHSSFPTLNLNPIFFYKFKSNKHNKPPFFSIRNCTTSNS 62
+R L SSSP++ LS SSFP+L + I + N P IR T S
Sbjct: 120 LRQLKLQPSSSPSS-----LSFSSSFPSLPIISISTTTLSPSNSNNPKGTFIRAAWTRRS 284
Query: 63 ISAKPSSQLRKNRSSNSDSDPKLTALRRLFSK 94
RK+ +D + L FSK
Sbjct: 285 RGEAEKKPSRKSWKRRTDMYMRPFLLDIFFSK 380
>BF647376 similar to GP|7939533|dbj| Nicotiana EREBP-3-like protein
{Arabidopsis thaliana}, partial (52%)
Length = 457
Score = 29.6 bits (65), Expect = 4.2
Identities = 14/41 (34%), Positives = 19/41 (46%)
Frame = +2
Query: 329 IEIRPYNSIVSEIENLAARGSSLWLDTSSVNAAIVNAYKAA 369
I RP+ +EI + +GS +WL T AY AA
Sbjct: 35 IRRRPWGKFAAEIRDPTRKGSRIWLGTFETAEQAARAYDAA 157
>TC76842 similar to GP|8809606|dbj|BAA97157.1 ethylene responsive element
binding factor 5 (ATERF5) {Arabidopsis thaliana},
partial (27%)
Length = 1135
Score = 29.6 bits (65), Expect = 4.2
Identities = 16/54 (29%), Positives = 23/54 (41%)
Frame = +1
Query: 319 EVDDHLKKANIEIRPYNSIVSEIENLAARGSSLWLDTSSVNAAIVNAYKAACDR 372
EV + + RP+ +EI + RGS +WL T AY +A R
Sbjct: 556 EVAEKQHYRGVRQRPWGKFAAEIRDPNKRGSRVWLGTFETAIEAAKAYDSAAFR 717
>CA919120 PIR|F84614|F8 probable kinesin heavy chain [imported] - Arabidopsis
thaliana, partial (1%)
Length = 661
Score = 29.6 bits (65), Expect = 4.2
Identities = 14/44 (31%), Positives = 22/44 (49%)
Frame = -1
Query: 54 IRNCTTSNSISAKPSSQLRKNRSSNSDSDPKLTALRRLFSKPDV 97
+R+ N++ PSS LR + + PK+ RL +KP V
Sbjct: 499 LRSNDIENNLVGPPSSTLRDRKMTRKSDPPKIVRTGRLTTKPPV 368
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.133 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,546,692
Number of Sequences: 36976
Number of extensions: 322095
Number of successful extensions: 1959
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 1910
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1942
length of query: 713
length of database: 9,014,727
effective HSP length: 103
effective length of query: 610
effective length of database: 5,206,199
effective search space: 3175781390
effective search space used: 3175781390
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146571.1


