

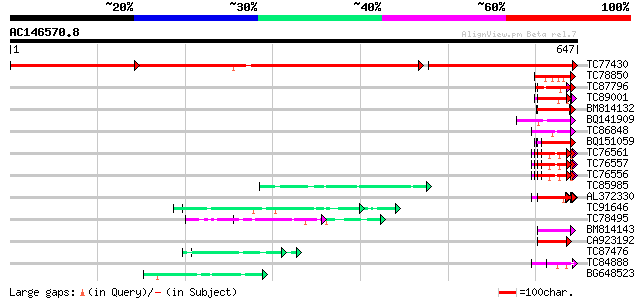
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146570.8 - phase: 0
(647 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77430 similar to GP|13676413|dbj|BAB41197. hypothetical protei... 597 0.0
TC78850 homologue to GP|10177413|dbj|BAB10544. fibrillarin-like ... 53 3e-07
TC87796 weakly similar to SP|P03211|EBN1_EBV EBNA-1 nuclear prot... 53 4e-07
TC89001 similar to GP|5726567|gb|AAD48471.1| glycine-rich RNA-bi... 51 2e-06
BM814132 weakly similar to OMNI|MT3615.1 PE_PGRS family protein ... 51 2e-06
BQ141909 weakly similar to GP|14253159|emb VMP3 protein {Volvox ... 50 4e-06
TC86848 similar to GP|10177413|dbj|BAB10544. fibrillarin-like {A... 50 4e-06
BQ151059 similar to GP|7291525|gb|A CG9888-PA {Drosophila melano... 49 5e-06
TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 49 6e-06
TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 49 6e-06
TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 49 6e-06
TC85985 similar to GP|5139695|dbj|BAA81686.1 expressed in cucumb... 49 8e-06
AL372330 similar to GP|21322752|dbj cold shock protein-1 {Tritic... 48 1e-05
TC91646 weakly similar to GP|6523547|emb|CAB62280.1 hydroxyproli... 48 1e-05
TC78495 homologue to PIR|S53504|S53504 extensin-like protein S3 ... 48 1e-05
BM814143 GP|21322711|e pherophorin-dz1 protein {Volvox carteri f... 47 2e-05
CA923192 similar to GP|20196906|gb| expressed protein {Arabidops... 46 5e-05
TC87476 homologue to PIR|T11622|T11622 extensin class 1 precurso... 45 7e-05
TC84888 similar to GP|21322752|dbj|BAB78536. cold shock protein-... 45 7e-05
BG648523 similar to GP|9955540|emb| putative protein {Arabidopsi... 45 9e-05
>TC77430 similar to GP|13676413|dbj|BAB41197. hypothetical protein {Glycine
max}, partial (71%)
Length = 2529
Score = 597 bits (1539), Expect(3) = 0.0
Identities = 303/335 (90%), Positives = 309/335 (91%), Gaps = 5/335 (1%)
Frame = +2
Query: 143 GSNFQGPLYQPGGNVVSWGASSPAPNANGGGLAMPMYWQGYYGAPNGLPQLHQQSLFRPP 202
G F+GPLYQPGGNVVSWGASSPAPNANGGGLAMPMYWQGYYGAPNGLPQLHQQSLFRPP
Sbjct: 524 GQTFKGPLYQPGGNVVSWGASSPAPNANGGGLAMPMYWQGYYGAPNGLPQLHQQSLFRPP 703
Query: 203 PGLSMPSSMQQPMPSSLQQPLPSSMQQPMPSSMQQPMPSSMQQPMPSSMQQP-----MLS 257
PGLSMPSSMQQPMPSSLQQPLPS+MQQPMPSSMQQPMPSSMQQPMPS +
Sbjct: 704 PGLSMPSSMQQPMPSSLQQPLPSAMQQPMPSSMQQPMPSSMQQPMPSLYAAADAFIYAAA 883
Query: 258 SMQQPLQFPSFSSSLPTGPSNFPEFPSAFLPVGTSSPNITSTSAPPLNLSTTILPPAPSA 317
P+ P + + GPSNFPEFPSAFLPVGTSSPNITSTSAPPLNLSTTILPPAPSA
Sbjct: 884 IAVSPVLVPHYQA----GPSNFPEFPSAFLPVGTSSPNITSTSAPPLNLSTTILPPAPSA 1051
Query: 318 TLAPETFQASVSNKAPTVSLPAATLGANLPSLAPFTSGGSDINAAVPLANKPNAISGSSL 377
TLAPETFQASVSNKAPTVSLPAATLGANLPSLAPFTSGGSDINAAVPLANKPNAISGSSL
Sbjct: 1052TLAPETFQASVSNKAPTVSLPAATLGANLPSLAPFTSGGSDINAAVPLANKPNAISGSSL 1231
Query: 378 SYQTVPQQMTPSIVGSSNSVRTEPPVPSLVTPAQLLQSGQTVAASSKPSQTPHKDVEVVQ 437
SYQTVPQQMTPSIVGSSNSVRTEPPVPSLVTPAQLLQSGQTVAASSKPSQTPHKDVEVVQ
Sbjct: 1232SYQTVPQQMTPSIVGSSNSVRTEPPVPSLVTPAQLLQSGQTVAASSKPSQTPHKDVEVVQ 1411
Query: 438 VSSTSAPEPSVPVAAESQPPILPLPATSRPIHRPG 472
VSSTSAPEPSVPVAAESQPPILPLPATSRPIHRPG
Sbjct: 1412VSSTSAPEPSVPVAAESQPPILPLPATSRPIHRPG 1516
Score = 371 bits (953), Expect(3) = 0.0
Identities = 168/169 (99%), Positives = 168/169 (99%)
Frame = +1
Query: 479 HHGYGYRGRGRGRGIGGFRPAERFTEDFDFTAMNEKFKKDEVWGHLGKSNKKDGEENASD 538
HHGYGYRGR RGRGIGGFRPAERFTEDFDFTAMNEKFKKDEVWGHLGKSNKKDGEENASD
Sbjct: 1537 HHGYGYRGRERGRGIGGFRPAERFTEDFDFTAMNEKFKKDEVWGHLGKSNKKDGEENASD 1716
Query: 539 EDGGQDEDNGDVSNLEVKPVYNKDDFFDSLSCNSLNHDPQNGRVRYSEQIKMDTETFGDF 598
EDGGQDEDNGDVSNLEVKPVYNKDDFFDSLSCNSLNHDPQNGRVRYSEQIKMDTETFGDF
Sbjct: 1717 EDGGQDEDNGDVSNLEVKPVYNKDDFFDSLSCNSLNHDPQNGRVRYSEQIKMDTETFGDF 1896
Query: 599 SRHRGGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGGRY 647
SRHRGGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGGRY
Sbjct: 1897 SRHRGGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGGRY 2043
Score = 295 bits (756), Expect(3) = 0.0
Identities = 148/148 (100%), Positives = 148/148 (100%)
Frame = +1
Query: 1 MASDTASRSSSAADSYIGCLISLTSKSEIRYEGVLYNINTDESSIGLKNVRSFGTEGRKK 60
MASDTASRSSSAADSYIGCLISLTSKSEIRYEGVLYNINTDESSIGLKNVRSFGTEGRKK
Sbjct: 97 MASDTASRSSSAADSYIGCLISLTSKSEIRYEGVLYNINTDESSIGLKNVRSFGTEGRKK 276
Query: 61 DGPQILPGDKVYEYILFRGTDIKDLQVKSSPPVQPAVPTNTDPAIIQSQYPRLATTSTSL 120
DGPQILPGDKVYEYILFRGTDIKDLQVKSSPPVQPAVPTNTDPAIIQSQYPRLATTSTSL
Sbjct: 277 DGPQILPGDKVYEYILFRGTDIKDLQVKSSPPVQPAVPTNTDPAIIQSQYPRLATTSTSL 456
Query: 121 PAVSGSLTDASPNPNTTQLGHPGSNFQG 148
PAVSGSLTDASPNPNTTQLGHPGSNFQG
Sbjct: 457 PAVSGSLTDASPNPNTTQLGHPGSNFQG 540
Score = 30.4 bits (67), Expect = 2.2
Identities = 14/26 (53%), Positives = 17/26 (64%)
Frame = +1
Query: 472 GGASNQTHHGYGYRGRGRGRGIGGFR 497
GG + + GYG RGRG GRG+ G R
Sbjct: 1963 GGYYGRGYGGYGGRGRGGGRGMPGGR 2040
>TC78850 homologue to GP|10177413|dbj|BAB10544. fibrillarin-like
{Arabidopsis thaliana}, partial (87%)
Length = 1177
Score = 53.1 bits (126), Expect = 3e-07
Identities = 36/64 (56%), Positives = 39/64 (60%), Gaps = 18/64 (28%)
Frame = +3
Query: 600 RHRGGWGGRG---PWRGGGGGR----ARGGYY-----GRGYGG-----YGGRGRGGGR-G 641
R RGG+GGRG RGGGGGR RGG + GRG+GG GGRGRGGGR G
Sbjct: 60 RGRGGFGGRGGDRGGRGGGGGRGFGGGRGGDFKPRGGGRGFGGGRGARGGGRGRGGGRGG 239
Query: 642 MPGG 645
M GG
Sbjct: 240 MKGG 251
Score = 41.2 bits (95), Expect = 0.001
Identities = 20/29 (68%), Positives = 21/29 (71%)
Frame = +3
Query: 618 RARGGYYGRGYGGYGGRGRGGGRGMPGGR 646
R RGG+ GRG G GGRG GGGRG GGR
Sbjct: 60 RGRGGFGGRG-GDRGGRGGGGGRGFGGGR 143
>TC87796 weakly similar to SP|P03211|EBN1_EBV EBNA-1 nuclear protein.
[strain B95-8 Human herpesvirus 4] {Epstein-barr
virus}, partial (23%)
Length = 431
Score = 52.8 bits (125), Expect = 4e-07
Identities = 26/45 (57%), Positives = 28/45 (61%)
Frame = +1
Query: 601 HRGGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
H GG+GG G GGGGG GG +G GYG GG G G G G PGG
Sbjct: 10 HGGGYGGGGG-SGGGGGGGAGGAHGVGYGSGGGTGGGYGGGSPGG 141
Score = 43.1 bits (100), Expect = 3e-04
Identities = 23/44 (52%), Positives = 25/44 (56%), Gaps = 5/44 (11%)
Frame = +1
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRG-----YGGYGGRGRGGGRG 641
GG+GG P GGGGG + GG G G YGG G G GGG G
Sbjct: 112 GGYGGGSPGGGGGGGGSGGGGGGGGAHGGAYGGGIGGGEGGGHG 243
Score = 38.9 bits (89), Expect = 0.006
Identities = 22/44 (50%), Positives = 23/44 (52%)
Frame = +1
Query: 595 FGDFSRHRGGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGG 638
+G S GG GG GGGGG GG YG G GG G G GG
Sbjct: 118 YGGGSPGGGGGGGGSGGGGGGGGA-HGGAYGGGIGGGEGGGHGG 246
Score = 31.2 bits (69), Expect = 1.3
Identities = 14/24 (58%), Positives = 15/24 (62%)
Frame = +1
Query: 621 GGYYGRGYGGYGGRGRGGGRGMPG 644
G +G GYGG GG G GGG G G
Sbjct: 1 GRDHGGGYGGGGGSGGGGGGGAGG 72
>TC89001 similar to GP|5726567|gb|AAD48471.1| glycine-rich RNA-binding
protein {Glycine max}, partial (90%)
Length = 488
Score = 50.8 bits (120), Expect = 2e-06
Identities = 26/43 (60%), Positives = 27/43 (62%), Gaps = 4/43 (9%)
Frame = +3
Query: 603 GGWGGRGPWRGGGGGRARGGYY----GRGYGGYGGRGRGGGRG 641
GG+GG G GGGGGR GGY G GYGG G RG GGG G
Sbjct: 342 GGYGGGGGGYGGGGGRRDGGYSRSGGGGGYGGGGDRGYGGGGG 470
Score = 46.6 bits (109), Expect = 3e-05
Identities = 30/56 (53%), Positives = 30/56 (53%), Gaps = 9/56 (16%)
Frame = +3
Query: 599 SRHRGGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRG---------RGGGRGMPGG 645
SR GG GGRG GGGGG GG YG G GGYGG G GGG G GG
Sbjct: 285 SRGSGG-GGRG---GGGGGYGGGGGYGGGGGGYGGGGGRRDGGYSRSGGGGGYGGG 440
Score = 45.4 bits (106), Expect = 7e-05
Identities = 24/44 (54%), Positives = 26/44 (58%)
Frame = +3
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGGR 646
GG+GG G + GGGGG GG GR GGY G GGG G G R
Sbjct: 324 GGYGGGGGYGGGGGGYGGGG--GRRDGGYSRSGGGGGYGGGGDR 449
Score = 39.3 bits (90), Expect = 0.005
Identities = 26/54 (48%), Positives = 26/54 (48%), Gaps = 11/54 (20%)
Frame = +3
Query: 603 GGWGGRGPWRGGGGGR--ARGGYYGRG---------YGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GGY G G GG GG G GG RG GG
Sbjct: 303 GGRGGGGGGYGGGGGYGGGGGGYGGGGGRRDGGYSRSGGGGGYGGGGDRGYGGG 464
Score = 33.9 bits (76), Expect = 0.20
Identities = 20/44 (45%), Positives = 21/44 (47%)
Frame = +3
Query: 596 GDFSRHRGGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGG 639
G R GG+ G G GGG RGYGG GG G GGG
Sbjct: 375 GGGGRRDGGYSRSGGGGGYGGGG------DRGYGGGGGGGYGGG 488
>BM814132 weakly similar to OMNI|MT3615.1 PE_PGRS family protein
{Mycobacterium tuberculosis CDC1551}, partial (7%)
Length = 164
Score = 50.8 bits (120), Expect = 2e-06
Identities = 27/44 (61%), Positives = 27/44 (61%)
Frame = +3
Query: 602 RGGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
RGG GG G GGGGG A GG G G GG GG G GGG G GG
Sbjct: 12 RGGGGGGGGGGGGGGGGAGGGGGGGGGGGGGGGGGGGGGGGGGG 143
Score = 50.1 bits (118), Expect = 3e-06
Identities = 26/43 (60%), Positives = 26/43 (60%)
Frame = +2
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG GRG GG GG G GGG G GG
Sbjct: 29 GGGGGGGGGGGGGGGGGGGGGGGRGGGGGGGGGGGGGGGGGGG 157
Score = 49.7 bits (117), Expect = 4e-06
Identities = 26/43 (60%), Positives = 26/43 (60%)
Frame = +2
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GGRG GGG G GG
Sbjct: 5 GGAGGGGGGGGGGGGGGGGGGGGGGGGGGGGRGGGGGGGGGGG 133
Score = 48.1 bits (113), Expect = 1e-05
Identities = 25/42 (59%), Positives = 25/42 (59%)
Frame = +1
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPG 644
GG GG G RGGGGG GG G G GG GG G GGG G G
Sbjct: 37 GGGGGGGGGRGGGGGGGGGGAGGGGGGGGGGGGGGGGGGGGG 162
Score = 47.8 bits (112), Expect = 1e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +2
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 23 GGGGGGGGGGGGGGGGGGGGGGGGGRGGGGGGGGGGGGGGGGG 151
Score = 47.8 bits (112), Expect = 1e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +3
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 36 GGGGGGGGGAGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 164
Score = 47.8 bits (112), Expect = 1e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +3
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 18 GGGGGGGGGGGGGGGAGGGGGGGGGGGGGGGGGGGGGGGGGGG 146
Score = 45.8 bits (107), Expect = 5e-05
Identities = 26/44 (59%), Positives = 26/44 (59%), Gaps = 1/44 (2%)
Frame = +1
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRG-YGGYGGRGRGGGRGMPGG 645
GG GG G GGGGGR GG G G GG GG G GGG G GG
Sbjct: 19 GGGGGGGGGGGGGGGRGGGGGGGGGGAGGGGGGGGGGGGGGGGG 150
Score = 45.4 bits (106), Expect = 7e-05
Identities = 24/43 (55%), Positives = 24/43 (55%)
Frame = +1
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG GGG G GG
Sbjct: 1 GGGGGGGGGGGGGGGGGGGGGRGGGGGGGGGGAGGGGGGGGGG 129
Score = 45.1 bits (105), Expect = 9e-05
Identities = 24/43 (55%), Positives = 24/43 (55%)
Frame = +3
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGG GG G G GG GG G GGG G GG
Sbjct: 33 GGGGGGGGGGAGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 161
Score = 37.0 bits (84), Expect = 0.024
Identities = 19/33 (57%), Positives = 19/33 (57%)
Frame = +2
Query: 614 GGGGRARGGYYGRGYGGYGGRGRGGGRGMPGGR 646
GGG GG G G GG GG G GGG G GGR
Sbjct: 2 GGGAGGGGGGGGGGGGGGGGGGGGGGGGGGGGR 100
Score = 36.6 bits (83), Expect = 0.031
Identities = 21/40 (52%), Positives = 21/40 (52%)
Frame = +1
Query: 602 RGGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRG 641
RGG GG G GGGG G G GG GG G GGG G
Sbjct: 64 RGGGGGGGGGGAGGGG-------GGGGGGGGGGGGGGGGG 162
>BQ141909 weakly similar to GP|14253159|emb VMP3 protein {Volvox carteri f.
nagariensis}, partial (13%)
Length = 1223
Score = 49.7 bits (117), Expect = 4e-06
Identities = 32/74 (43%), Positives = 40/74 (53%), Gaps = 7/74 (9%)
Frame = +3
Query: 579 NGRVRYSEQIKMDTETFGDFSRH-----RGGWGGRGPWRGGGGGRARGGYYGR--GYGGY 631
NG+++ +E+I+ E G+ +GG GGRG G GGG RGG G G GG
Sbjct: 90 NGKIKTTEEIQRRKEGGGEEVVR*EEGVKGGSGGRG---GRGGGERRGGRRGEEGGGGGG 260
Query: 632 GGRGRGGGRGMPGG 645
GG G GGG G GG
Sbjct: 261 GGEGGGGGGGRKGG 302
Score = 41.6 bits (96), Expect = 0.001
Identities = 23/47 (48%), Positives = 25/47 (52%)
Frame = +2
Query: 600 RHRGGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGGR 646
R R G GGRG R G G + GG GRG GG GGG+G GR
Sbjct: 239 RGRRGGGGRGGGRRGRGKKGGGGTKGRGKRERGGGTEGGGKGKIEGR 379
Score = 34.3 bits (77), Expect = 0.15
Identities = 22/47 (46%), Positives = 23/47 (48%), Gaps = 2/47 (4%)
Frame = +2
Query: 602 RGGWGGRGPWRGGGGGRARGGYYGRGY--GGYGGRGRGGGRGMPGGR 646
RG G+ RG G R GG G G G GRGR GG G GGR
Sbjct: 137 RGRGSGKMRRRGERGKRGEGGEGGGGAEGGSERGRGRRGGGGRGGGR 277
Score = 34.3 bits (77), Expect = 0.15
Identities = 26/54 (48%), Positives = 26/54 (48%), Gaps = 14/54 (25%)
Frame = +2
Query: 600 RHRGGWGG--------RGPWRGGGGGRARGGYYGRGYGGYGG------RGRGGG 639
R GG GG RG R GGGGR GG GRG G GG R RGGG
Sbjct: 185 RGEGGEGGGGAEGGSERGRGRRGGGGRG-GGRRGRGKKGGGGTKGRGKRERGGG 343
Score = 32.3 bits (72), Expect = 0.58
Identities = 23/63 (36%), Positives = 32/63 (50%), Gaps = 3/63 (4%)
Frame = -1
Query: 262 PLQFPSFSSSLPTGPSNFPEFPSAFLPVGTSSPNITSTSAPPLNL---STTILPPAPSAT 318
PL FP SS + P FP P FLP+ +SP + PPL+ S+ + P P ++
Sbjct: 416 PLHFPPASSLSHSFPLFFPCRPPPFLPLSHASPFL--LFLPPLSSPSPSSPLPSPPPPSS 243
Query: 319 LAP 321
L P
Sbjct: 242 LFP 234
Score = 32.0 bits (71), Expect = 0.76
Identities = 29/99 (29%), Positives = 36/99 (36%), Gaps = 11/99 (11%)
Frame = -3
Query: 186 APNGLPQLHQQSLFRPPPGLSMPSSMQQPMP----------SSLQQPLPSSMQQPMPSSM 235
+P LP S+F PP P + P+P L PLP P+P S
Sbjct: 405 SPRLLPVPFLPSIFPLPPPSVPPPLSRFPLPFVPPPPFFPLPLLPPPLPPPPLLPLPLSD 226
Query: 236 QQPMPSSMQQPMPSSMQQPMLSSMQQPLQ-FPSFSSSLP 273
P P P P+L + PL PSF LP
Sbjct: 225 PPSAPPPPSPPSPRFPLSPLLLILPLPLPLLPSFVVFLP 109
Score = 29.6 bits (65), Expect = 3.8
Identities = 22/57 (38%), Positives = 23/57 (39%), Gaps = 14/57 (24%)
Frame = +3
Query: 603 GGWGGRGPWRGGGGGR------ARGGYYGRGYGGYG--------GRGRGGGRGMPGG 645
GG GG G GGGGGR GG G G G G+GGG G G
Sbjct: 246 GGGGGGGEGGGGGGGRKGGEEQKEGGSVREGEERRGAAREK*REGMGQGGGGGKVEG 416
>TC86848 similar to GP|10177413|dbj|BAB10544. fibrillarin-like {Arabidopsis
thaliana}, partial (92%)
Length = 1250
Score = 49.7 bits (117), Expect = 4e-06
Identities = 37/64 (57%), Positives = 37/64 (57%), Gaps = 14/64 (21%)
Frame = +3
Query: 596 GDFSRHRGG-WGGRGPWRGGGGGR---------ARGGYYGRGYGGYGGR--GRGGGR--G 641
G F RGG GGRG RGG GGR ARGG GRG GG GGR GRGGGR G
Sbjct: 87 GGFRGGRGGDRGGRGGGRGGFGGRGGDRGTPFKARGG--GRGGGGRGGRGGGRGGGRGGG 260
Query: 642 MPGG 645
M GG
Sbjct: 261 MKGG 272
Score = 37.7 bits (86), Expect = 0.014
Identities = 26/52 (50%), Positives = 27/52 (51%), Gaps = 15/52 (28%)
Frame = +3
Query: 610 PWRGGGGGRARGGY------YGRGYGGYGGRG---------RGGGRGMPGGR 646
P RG GGG RGG G G GG+GGRG RGGGRG GGR
Sbjct: 66 PVRGRGGGGFRGGRGGDRGGRGGGRGGFGGRGGDRGTPFKARGGGRG-GGGR 218
Score = 37.0 bits (84), Expect = 0.024
Identities = 26/52 (50%), Positives = 27/52 (51%), Gaps = 5/52 (9%)
Frame = +3
Query: 600 RHRGGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMP-----GGR 646
R RGG G RG G GGR G G GG+GGRG G RG P GGR
Sbjct: 72 RGRGGGGFRGGRGGDRGGR------GGGRGGFGGRG--GDRGTPFKARGGGR 203
>BQ151059 similar to GP|7291525|gb|A CG9888-PA {Drosophila melanogaster},
partial (26%)
Length = 308
Score = 49.3 bits (116), Expect = 5e-06
Identities = 26/44 (59%), Positives = 26/44 (59%)
Frame = +1
Query: 602 RGGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
RGG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 34 RGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 165
Score = 47.8 bits (112), Expect = 1e-05
Identities = 26/46 (56%), Positives = 26/46 (56%)
Frame = +1
Query: 600 RHRGGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
R GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 34 RGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 171
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +3
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 75 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 203
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +2
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 53 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 181
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +1
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 46 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 174
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +2
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 59 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 187
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +2
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 65 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 193
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +2
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 89 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 217
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +1
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 67 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 195
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +3
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 72 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 200
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +2
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 56 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 184
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +1
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 76 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 204
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +1
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 79 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 207
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +1
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 82 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 210
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +2
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 137 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 265
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +3
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 135 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 263
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +2
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 92 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 220
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +1
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 94 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 222
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +2
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 98 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 226
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +1
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 100 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 228
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +3
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 105 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 233
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +2
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 107 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 235
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +3
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 111 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 239
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +3
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 114 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 242
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +2
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 116 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 244
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +3
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 120 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 248
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +2
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 122 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 250
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +1
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 124 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 252
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +1
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 127 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 255
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +2
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 131 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 259
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +1
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 85 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 213
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +1
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 61 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 189
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +2
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 140 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 268
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +3
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 144 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 272
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +2
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 146 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 274
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +1
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 148 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 276
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +1
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 151 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 279
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +2
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 155 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 283
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +3
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 159 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 287
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +3
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 162 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 290
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +2
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 164 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 292
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +1
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 166 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 294
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +1
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 169 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 297
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +1
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 172 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 300
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +3
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 177 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 305
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +3
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 180 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 308
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +3
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 51 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 179
Score = 43.9 bits (102), Expect = 2e-04
Identities = 23/38 (60%), Positives = 23/38 (60%)
Frame = +1
Query: 608 RGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
RG RGGGGG GG G G GG GG G GGG G GG
Sbjct: 22 RGKPRGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 135
Score = 43.1 bits (100), Expect = 3e-04
Identities = 22/39 (56%), Positives = 22/39 (56%)
Frame = +2
Query: 607 GRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
G P GGGGG GG G G GG GG G GGG G GG
Sbjct: 23 GENPGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 139
>TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 1054
Score = 48.9 bits (115), Expect = 6e-06
Identities = 26/46 (56%), Positives = 28/46 (60%), Gaps = 3/46 (6%)
Frame = +1
Query: 599 SRHRGGWGGRGPWRGGGGGRARGGYYG---RGYGGYGGRGRGGGRG 641
++ RG GG G RGGGG GG YG RGYGG GG G GGG G
Sbjct: 583 AQSRGSGGGGGGGRGGGGYGGGGGGYGGERRGYGGGGGYGGGGGGG 720
Score = 47.8 bits (112), Expect = 1e-05
Identities = 26/39 (66%), Positives = 27/39 (68%)
Frame = +1
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRG 641
GG GG G RGGGGG +RGG G GYGG GG RGGG G
Sbjct: 706 GGGGGYGERRGGGGGYSRGGG-GGGYGG-GGYSRGGGDG 816
Score = 47.4 bits (111), Expect = 2e-05
Identities = 26/40 (65%), Positives = 26/40 (65%), Gaps = 2/40 (5%)
Frame = +1
Query: 608 RGPWRGGGGGRARGGYYGRGYGGYGG--RGRGGGRGMPGG 645
RG GGGGGR GG YG G GGYGG RG GGG G GG
Sbjct: 592 RGSGGGGGGGRGGGG-YGGGGGGYGGERRGYGGGGGYGGG 708
Score = 45.4 bits (106), Expect = 7e-05
Identities = 26/52 (50%), Positives = 28/52 (53%), Gaps = 2/52 (3%)
Frame = +1
Query: 596 GDFSRHRGGWGG--RGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
G + GG+GG RG GGG G GG YG GG GG RGGG G GG
Sbjct: 631 GGYGGGGGGYGGERRGYGGGGGYGGGGGGGYGERRGGGGGYSRGGGGGGYGG 786
Score = 43.5 bits (101), Expect = 3e-04
Identities = 30/60 (50%), Positives = 31/60 (51%), Gaps = 8/60 (13%)
Frame = +1
Query: 596 GDFSRHRGGWGGRGPWRGG-----GGGRARGGYYGRGYGGYGGRGRGGG---RGMPGGRY 647
G R GG+GG G GG GGG GGY G G GGYG R GGG RG GG Y
Sbjct: 610 GGGGRGGGGYGGGGGGYGGERRGYGGG---GGYGGGGGGGYGERRGGGGGYSRGGGGGGY 780
Score = 41.2 bits (95), Expect = 0.001
Identities = 25/53 (47%), Positives = 28/53 (52%), Gaps = 3/53 (5%)
Frame = +1
Query: 596 GDFSRHRGGWGGRGPWRGGGGG---RARGGYYGRGYGGYGGRGRGGGRGMPGG 645
G + R G+GG G + GGGGG RGG G GG GG G GGG GG
Sbjct: 652 GGYGGERRGYGGGGGYGGGGGGGYGERRGGGGGYSRGG-GGGGYGGGGYSRGG 807
Score = 37.4 bits (85), Expect = 0.018
Identities = 21/38 (55%), Positives = 22/38 (57%)
Frame = +1
Query: 596 GDFSRHRGGWGGRGPWRGGGGGRARGGYYGRGYGGYGG 633
G + RGG GG RGGGGG GG Y RG GG GG
Sbjct: 715 GGYGERRGGGGGYS--RGGGGGGYGGGGYSRG-GGDGG 819
>TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 912
Score = 48.9 bits (115), Expect = 6e-06
Identities = 26/46 (56%), Positives = 28/46 (60%), Gaps = 3/46 (6%)
Frame = +1
Query: 599 SRHRGGWGGRGPWRGGGGGRARGGYYG---RGYGGYGGRGRGGGRG 641
++ RG GG G RGGGG GG YG RGYGG GG G GGG G
Sbjct: 487 AQSRGSGGGGGGGRGGGGYGGGGGGYGGERRGYGGGGGYGGGGGGG 624
Score = 47.8 bits (112), Expect = 1e-05
Identities = 26/39 (66%), Positives = 27/39 (68%)
Frame = +1
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRG 641
GG GG G RGGGGG +RGG G GYGG GG RGGG G
Sbjct: 610 GGGGGYGERRGGGGGYSRGGG-GGGYGG-GGYSRGGGDG 720
Score = 47.4 bits (111), Expect = 2e-05
Identities = 26/40 (65%), Positives = 26/40 (65%), Gaps = 2/40 (5%)
Frame = +1
Query: 608 RGPWRGGGGGRARGGYYGRGYGGYGG--RGRGGGRGMPGG 645
RG GGGGGR GG YG G GGYGG RG GGG G GG
Sbjct: 496 RGSGGGGGGGRGGGG-YGGGGGGYGGERRGYGGGGGYGGG 612
Score = 45.4 bits (106), Expect = 7e-05
Identities = 26/52 (50%), Positives = 28/52 (53%), Gaps = 2/52 (3%)
Frame = +1
Query: 596 GDFSRHRGGWGG--RGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
G + GG+GG RG GGG G GG YG GG GG RGGG G GG
Sbjct: 535 GGYGGGGGGYGGERRGYGGGGGYGGGGGGGYGERRGGGGGYSRGGGGGGYGG 690
Score = 43.5 bits (101), Expect = 3e-04
Identities = 30/60 (50%), Positives = 31/60 (51%), Gaps = 8/60 (13%)
Frame = +1
Query: 596 GDFSRHRGGWGGRGPWRGG-----GGGRARGGYYGRGYGGYGGRGRGGG---RGMPGGRY 647
G R GG+GG G GG GGG GGY G G GGYG R GGG RG GG Y
Sbjct: 514 GGGGRGGGGYGGGGGGYGGERRGYGGG---GGYGGGGGGGYGERRGGGGGYSRGGGGGGY 684
Score = 41.2 bits (95), Expect = 0.001
Identities = 25/53 (47%), Positives = 28/53 (52%), Gaps = 3/53 (5%)
Frame = +1
Query: 596 GDFSRHRGGWGGRGPWRGGGGG---RARGGYYGRGYGGYGGRGRGGGRGMPGG 645
G + R G+GG G + GGGGG RGG G GG GG G GGG GG
Sbjct: 556 GGYGGERRGYGGGGGYGGGGGGGYGERRGGGGGYSRGG-GGGGYGGGGYSRGG 711
Score = 37.4 bits (85), Expect = 0.018
Identities = 21/38 (55%), Positives = 22/38 (57%)
Frame = +1
Query: 596 GDFSRHRGGWGGRGPWRGGGGGRARGGYYGRGYGGYGG 633
G + RGG GG RGGGGG GG Y RG GG GG
Sbjct: 619 GGYGERRGGGGGYS--RGGGGGGYGGGGYSRG-GGDGG 723
>TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 754
Score = 48.9 bits (115), Expect = 6e-06
Identities = 26/46 (56%), Positives = 28/46 (60%), Gaps = 3/46 (6%)
Frame = +2
Query: 599 SRHRGGWGGRGPWRGGGGGRARGGYYG---RGYGGYGGRGRGGGRG 641
++ RG GG G RGGGG GG YG RGYGG GG G GGG G
Sbjct: 329 AQSRGSGGGGGGGRGGGGYGGGGGGYGGERRGYGGGGGYGGGGGGG 466
Score = 47.8 bits (112), Expect = 1e-05
Identities = 26/39 (66%), Positives = 27/39 (68%)
Frame = +2
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRG 641
GG GG G RGGGGG +RGG G GYGG GG RGGG G
Sbjct: 452 GGGGGYGERRGGGGGYSRGGG-GGGYGG-GGYSRGGGDG 562
Score = 47.4 bits (111), Expect = 2e-05
Identities = 26/40 (65%), Positives = 26/40 (65%), Gaps = 2/40 (5%)
Frame = +2
Query: 608 RGPWRGGGGGRARGGYYGRGYGGYGG--RGRGGGRGMPGG 645
RG GGGGGR GG YG G GGYGG RG GGG G GG
Sbjct: 338 RGSGGGGGGGRGGGG-YGGGGGGYGGERRGYGGGGGYGGG 454
Score = 45.4 bits (106), Expect = 7e-05
Identities = 26/52 (50%), Positives = 28/52 (53%), Gaps = 2/52 (3%)
Frame = +2
Query: 596 GDFSRHRGGWGG--RGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
G + GG+GG RG GGG G GG YG GG GG RGGG G GG
Sbjct: 377 GGYGGGGGGYGGERRGYGGGGGYGGGGGGGYGERRGGGGGYSRGGGGGGYGG 532
Score = 43.5 bits (101), Expect = 3e-04
Identities = 30/60 (50%), Positives = 31/60 (51%), Gaps = 8/60 (13%)
Frame = +2
Query: 596 GDFSRHRGGWGGRGPWRGG-----GGGRARGGYYGRGYGGYGGRGRGGG---RGMPGGRY 647
G R GG+GG G GG GGG GGY G G GGYG R GGG RG GG Y
Sbjct: 356 GGGGRGGGGYGGGGGGYGGERRGYGGG---GGYGGGGGGGYGERRGGGGGYSRGGGGGGY 526
Score = 41.2 bits (95), Expect = 0.001
Identities = 25/53 (47%), Positives = 28/53 (52%), Gaps = 3/53 (5%)
Frame = +2
Query: 596 GDFSRHRGGWGGRGPWRGGGGG---RARGGYYGRGYGGYGGRGRGGGRGMPGG 645
G + R G+GG G + GGGGG RGG G GG GG G GGG GG
Sbjct: 398 GGYGGERRGYGGGGGYGGGGGGGYGERRGGGGGYSRGG-GGGGYGGGGYSRGG 553
Score = 37.4 bits (85), Expect = 0.018
Identities = 21/38 (55%), Positives = 22/38 (57%)
Frame = +2
Query: 596 GDFSRHRGGWGGRGPWRGGGGGRARGGYYGRGYGGYGG 633
G + RGG GG RGGGGG GG Y RG GG GG
Sbjct: 461 GGYGERRGGGGGYS--RGGGGGGYGGGGYSRG-GGDGG 565
>TC85985 similar to GP|5139695|dbj|BAA81686.1 expressed in cucumber
hypocotyls {Cucumis sativus}, partial (42%)
Length = 892
Score = 48.5 bits (114), Expect = 8e-06
Identities = 52/197 (26%), Positives = 77/197 (38%), Gaps = 1/197 (0%)
Frame = +3
Query: 286 FLPVGTSSPNITSTSAPPLNLSTTILPPAPSATLAPETFQASVSNKAPTVSLPAATLGAN 345
F VG SP+ T++PP T+ P+ + AP ++ +P S PA++
Sbjct: 102 FAGVGGQSPSSAPTTSPP-----TVTTPSAAPVAAPTKPKSPAPVASPKSSPPASS---- 254
Query: 346 LPSLAPFTSGGSDINAAVPLANKPNAISGSSLSYQTVPQQMTPSIVGSSNSVRTEPPVPS 405
P+ A T S A VP+A P A S T P+ S + V + PP P
Sbjct: 255 -PTAATVTPAVSPA-APVPVAKSPAASSPVVAPVSTPPKPAPVSSPPAPVPV-SSPPTPV 425
Query: 406 LVTPAQLLQSGQTVAASSKPSQTPHKDVEVVQVSST-SAPEPSVPVAAESQPPILPLPAT 464
V+ + ++ P+ P K + + SAP PS + PP+ P
Sbjct: 426 PVSSPPTASTPAVTPSAEVPAAAPSKSKKKTKKGKKHSAPAPSPALEGPPAPPV-GAPGP 602
Query: 465 SRPIHRPGGASNQTHHG 481
S PG AS G
Sbjct: 603 SLDASSPGPASAADESG 653
>AL372330 similar to GP|21322752|dbj cold shock protein-1 {Triticum
aestivum}, partial (33%)
Length = 435
Score = 47.8 bits (112), Expect = 1e-05
Identities = 25/50 (50%), Positives = 27/50 (54%)
Frame = +2
Query: 596 GDFSRHRGGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
G + GG+GG G G GG GGY G GGYGG G GGG G GG
Sbjct: 143 GGYGGSSGGYGGGGYGGGASGGYGGGGYGGGSGGGYGGGGYGGG-GYGGG 289
Score = 47.8 bits (112), Expect = 1e-05
Identities = 22/38 (57%), Positives = 24/38 (62%)
Frame = +2
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGR 640
GG+GG G G GGG GGY G GYGG GG G GG +
Sbjct: 203 GGYGGGGYGGGSGGGYGGGGYGGGGYGGGGGGGYGGDK 316
Score = 46.2 bits (108), Expect = 4e-05
Identities = 27/58 (46%), Positives = 29/58 (49%), Gaps = 12/58 (20%)
Frame = +2
Query: 596 GDFSRHRGGWGGRGPWRGGGG--GRARGGYYGRGYGG----------YGGRGRGGGRG 641
G + GG+GG GGGG G A GGY G GYGG YGG G GGG G
Sbjct: 122 GGYGGSSGGYGGSSGGYGGGGYGGGASGGYGGGGYGGGSGGGYGGGGYGGGGYGGGGG 295
Score = 45.4 bits (106), Expect = 7e-05
Identities = 25/48 (52%), Positives = 27/48 (56%), Gaps = 3/48 (6%)
Frame = +2
Query: 603 GGWGGR-GPWRGGGGGRARGGYYGRGYGGYGGRGRGGGR--GMPGGRY 647
GG+GG G + G GG GGY G GGYGG G GGG G GG Y
Sbjct: 122 GGYGGSSGGYGGSSGGYGGGGYGGGASGGYGGGGYGGGSGGGYGGGGY 265
Score = 44.7 bits (104), Expect = 1e-04
Identities = 26/47 (55%), Positives = 30/47 (63%), Gaps = 3/47 (6%)
Frame = +2
Query: 603 GGWGG--RGPWRGGG-GGRARGGYYGRGYGGYGGRGRGGGRGMPGGR 646
GG+GG G + GGG GG + GGY G GYGG GG G GGG G G +
Sbjct: 179 GGYGGGASGGYGGGGYGGGSGGGYGGGGYGG-GGYGGGGGGGYGGDK 316
Score = 39.3 bits (90), Expect = 0.005
Identities = 19/35 (54%), Positives = 20/35 (56%)
Frame = +2
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRG 637
GG+GG GGGG GGY G G GGYGG G
Sbjct: 218 GGYGGGSGGGYGGGGYGGGGYGGGGGGGYGGDKMG 322
Score = 35.4 bits (80), Expect = 0.069
Identities = 21/49 (42%), Positives = 22/49 (44%), Gaps = 6/49 (12%)
Frame = +2
Query: 603 GGWGGRGPWR------GGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG+GG GGG G G YG GGYGG G G G GG
Sbjct: 47 GGYGGSSSSSYPASSSGGGYGGGSSGGYGGSSGGYGGSSGGYGGGGYGG 193
Score = 31.6 bits (70), Expect = 0.99
Identities = 18/42 (42%), Positives = 19/42 (44%), Gaps = 9/42 (21%)
Frame = +2
Query: 613 GGGGGRARGGYYGRGY---------GGYGGRGRGGGRGMPGG 645
GG GG + GGY G GGYGG GG G GG
Sbjct: 23 GGYGGSSSGGYGGSSSSSYPASSSGGGYGGGSSGGYGGSSGG 148
Score = 30.8 bits (68), Expect = 1.7
Identities = 15/30 (50%), Positives = 16/30 (53%)
Frame = +2
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYG 632
GG+GG G GG GG GGY G G G
Sbjct: 242 GGYGGGGYGGGGYGGGGGGGYGGDKMGALG 331
>TC91646 weakly similar to GP|6523547|emb|CAB62280.1 hydroxyproline-rich
glycoprotein DZ-HRGP {Volvox carteri f. nagariensis},
partial (15%)
Length = 984
Score = 47.8 bits (112), Expect = 1e-05
Identities = 71/259 (27%), Positives = 99/259 (37%), Gaps = 10/259 (3%)
Frame = +1
Query: 198 LFRPPPGLSMPSSMQQPMPSSLQQPLPSSMQQPMPSSMQQPMPSSMQQPMPSSMQQPMLS 257
L PP + PS P P S +P P S + + S +P+P S P P P LS
Sbjct: 310 LTTPPSLTTSPSPTASPPPRSPPRPPPRSPPRQLMSP-PRPLPRS---PSPPPRNPPELS 477
Query: 258 SMQQPLQFPSFSSSLPTGP-------SNFPEFPSA--FLPVGTSSPNITSTSAPPLNLST 308
+ P S PT P S P P+ LP P + S+ A L L+
Sbjct: 478 RTTPTPRPPMSPSPSPTSPPAATPPVSRPPPRPALRRLLPPPPLPPTLPSSMAS-LFLAA 654
Query: 309 TILPPAPSATLAPETFQASVSNKAPTVSLPA-ATLGANLPSLAPFTSGGSDINAAVPLAN 367
++LPPA T S+ A + LPA ATL + + +P TS +
Sbjct: 655 SVLPPASPPTT*SSPPTP*PSSAALLLCLPASATLPCSEQTASPVTSWTTSTRC------ 816
Query: 368 KPNAISGSSLSYQTVPQQMTPSIVGSSNSVRTEPPVPSLVTPAQLLQSGQTVAASSKPSQ 427
P+A + ++LS T P +V P LL S V++S +PS
Sbjct: 817 -PSARTSATLSAPV-----------------TLPSAAVVVLP--LLSS---VSSSPRPSA 927
Query: 428 TPHKDVEVVQVSSTSAPEP 446
+P TS P P
Sbjct: 928 SPSTSASTAPSVPTSPPSP 984
Score = 42.4 bits (98), Expect = 6e-04
Identities = 52/221 (23%), Positives = 84/221 (37%), Gaps = 3/221 (1%)
Frame = +1
Query: 187 PNGLPQLHQQSLFRPPPGLSMPSSMQQPMPSSLQQPLPSSMQQPMPSSMQQPMPSSMQQP 246
P P+ + L PP L S P L + P+ P P P P+S
Sbjct: 376 PRPPPRSPPRQLMSPPRPLPRSPSPPPRNPPELSRTTPT----PRPPMSPSPSPTSPPAA 543
Query: 247 MPSSMQQPMLSSMQQPLQFPSFSSSLPTGPSNFPEFPSAFLPVGTSSPNITSTSAP---P 303
P + P ++++ L P +LP+ ++ S P +SP T +S P P
Sbjct: 544 TPPVSRPPPRPALRRLLPPPPLPPTLPSSMASLFLAASVLPP---ASPPTT*SSPPTP*P 714
Query: 304 LNLSTTILPPAPSATLAPETFQASVSNKAPTVSLPAATLGANLPSLAPFTSGGSDINAAV 363
+ + + PA + E + V++ + P+A A L AP T + + +
Sbjct: 715 SSAALLLCLPASATLPCSEQTASPVTSWTTSTRCPSARTSATLS--APVTLPSAAV-VVL 885
Query: 364 PLANKPNAISGSSLSYQTVPQQMTPSIVGSSNSVRTEPPVP 404
PL SS+S P + ++ SV T PP P
Sbjct: 886 PLL--------SSVSSSPRPSASPSTSASTAPSVPTSPPSP 984
Score = 33.1 bits (74), Expect = 0.34
Identities = 29/97 (29%), Positives = 39/97 (39%)
Frame = +1
Query: 375 SSLSYQTVPQQMTPSIVGSSNSVRTEPPVPSLVTPAQLLQSGQTVAASSKPSQTPHKDVE 434
S L T P T +S R+ P P P QL+ + + S PS P E
Sbjct: 298 SRLPLTTPPSLTTSPSPTASPPPRSPPRPPPRSPPRQLMSPPRPLPRS--PSPPPRNPPE 471
Query: 435 VVQVSSTSAPEPSVPVAAESQPPILPLPATSRPIHRP 471
+ + + T P P P + + PP P SRP RP
Sbjct: 472 LSRTTPTPRP-PMSPSPSPTSPPAAT-PPVSRPPPRP 576
>TC78495 homologue to PIR|S53504|S53504 extensin-like protein S3 - alfalfa,
partial (97%)
Length = 1049
Score = 47.8 bits (112), Expect = 1e-05
Identities = 54/169 (31%), Positives = 70/169 (40%), Gaps = 8/169 (4%)
Frame = +3
Query: 201 PPPGLSMPSSMQQPMPSSLQQPLPSSMQQPMPSSMQQPMPSSMQQPMPSSMQQPMLSSMQ 260
PPP S P +Q P PL SS P P+ Q P +Q P Q P + +
Sbjct: 321 PPPVQSSPPPVQSSPP-----PLQSS---PPPA---QSTPPPVQSSPPPVQQSPPPTPLT 467
Query: 261 QPLQFPSFSSSLPTGPSNFPEFPSAFLPVGTSSPNITSTSA-PPLNLSTTILPPAPSATL 319
P P S+ P P P P F P + P T A PP L+ T L P+ T
Sbjct: 468 PP---PVQSTPPPASPP--PASPPPFSPPPATPPPATPPPATPPPALTPTPLSSPPATTP 632
Query: 320 APETFQASVSNKAPTVS-------LPAATLGANLPSLAPFTSGGSDINA 361
AP A + +KAP ++ PA L + PS++P SG D A
Sbjct: 633 APA--PAKLKSKAPALAPVLSPSDAPAPGLSSLSPSISP--SGTDDSGA 767
Score = 44.3 bits (103), Expect = 1e-04
Identities = 47/178 (26%), Positives = 66/178 (36%), Gaps = 4/178 (2%)
Frame = +3
Query: 256 LSSMQQPLQFPSFSSSLPTGPSNF-PEFPSAFLPVGTSSPNITSTSAPPLNLSTTILPPA 314
+ + Q P P+ S + PT P+N P P A P SSP +S PPL S PPA
Sbjct: 225 VGAQQAPSTSPNSSPAPPTPPANTPPTTPQASPPPVQSSPPPVQSSPPPLQSSP---PPA 395
Query: 315 PSATLAPETFQASVSNKAPTVSLPAATLGANLPSLAPFTSGGSDIN---AAVPLANKPNA 371
S ++ V P L + + P +P + + A P A P A
Sbjct: 396 QSTPPPVQSSPPPVQQSPPPTPLTPPPVQSTPPPASPPPASPPPFSPPPATPPPATPPPA 575
Query: 372 ISGSSLSYQTVPQQMTPSIVGSSNSVRTEPPVPSLVTPAQLLQSGQTVAASSKPSQTP 429
T P +TP+ + S + P PA+L +A PS P
Sbjct: 576 ---------TPPPALTPTPLSSPPATTPAP------APAKLKSKAPALAPVLSPSDAP 704
Score = 38.5 bits (88), Expect = 0.008
Identities = 38/143 (26%), Positives = 53/143 (36%)
Frame = +3
Query: 332 APTVSLPAATLGANLPSLAPFTSGGSDINAAVPLANKPNAISGSSLSYQTVPQQMTPSIV 391
+P S T AN P P S ++ P+ + P + S P Q TP V
Sbjct: 252 SPNSSPAPPTPPANTPPTTPQASPPPVQSSPPPVQSSPPPLQSSP-----PPAQSTPPPV 416
Query: 392 GSSNSVRTEPPVPSLVTPAQLLQSGQTVAASSKPSQTPHKDVEVVQVSSTSAPEPSVPVA 451
SS + P P+ +TP + QS A+ S P + P P+ P
Sbjct: 417 QSSPPPVQQSPPPTPLTPPPV-QSTPPPASPPPASPPPFSP-------PPATPPPATPPP 572
Query: 452 AESQPPILPLPATSRPIHRPGGA 474
A P + P P +S P P A
Sbjct: 573 ATPPPALTPTPLSSPPATTPAPA 641
Score = 35.4 bits (80), Expect = 0.069
Identities = 46/188 (24%), Positives = 66/188 (34%), Gaps = 5/188 (2%)
Frame = +3
Query: 89 SSPPVQPAVPTNTDPAIIQSQYPRLATT----STSLPAVSGSLTDASPNPNTTQLGHPGS 144
+S P P P NT P Q+ P + ++ +S P + S A P Q P
Sbjct: 258 NSSPAPPTPPANTPPTTPQASPPPVQSSPPPVQSSPPPLQSSPPPAQSTPPPVQSSPPPV 437
Query: 145 NFQGPLYQPGGNVVSWGASSPAPNANGGGLAMPMYWQGYYGAPNGLPQLHQQSLFRPPPG 204
Q P P +P P + A P P P PPP
Sbjct: 438 Q-QSPPPTP---------LTPPPVQSTPPPASP---------PPASPPPFSPPPATPPPA 560
Query: 205 LSMPSSMQQPM-PSSLQQPLPSSMQQPMPSSMQQPMPSSMQQPMPSSMQQPMLSSMQQPL 263
P++ + P+ L P P++ P P+ ++ P+ PS P LSS+
Sbjct: 561 TPPPATPPPALTPTPLSSP-PATTPAPAPAKLKSKAPALAPVLSPSDAPAPGLSSLS--- 728
Query: 264 QFPSFSSS 271
PS S S
Sbjct: 729 --PSISPS 746
Score = 34.7 bits (78), Expect = 0.12
Identities = 34/126 (26%), Positives = 43/126 (33%)
Frame = +3
Query: 350 APFTSGGSDINAAVPLANKPNAISGSSLSYQTVPQQMTPSIVGSSNSVRTEPPVPSLVTP 409
AP TS S P AN P T PQ P + S V++ PP P +P
Sbjct: 240 APSTSPNSSPAPPTPPANTP----------PTTPQASPPPVQSSPPPVQSSPP-PLQSSP 386
Query: 410 AQLLQSGQTVAASSKPSQTPHKDVEVVQVSSTSAPEPSVPVAAESQPPILPLPATSRPIH 469
+ V +S P V + P P P +S PP P S P
Sbjct: 387 PPAQSTPPPVQSSPPP------------VQQSPPPTPLTPPPVQSTPPPASPPPASPPPF 530
Query: 470 RPGGAS 475
P A+
Sbjct: 531 SPPPAT 548
>BM814143 GP|21322711|e pherophorin-dz1 protein {Volvox carteri f.
nagariensis}, partial (4%)
Length = 132
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +1
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 4 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 132
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/43 (58%), Positives = 25/43 (58%)
Frame = +3
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG GG G GGGGG GG G G GG GG G GGG G GG
Sbjct: 3 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 131
>CA923192 similar to GP|20196906|gb| expressed protein {Arabidopsis
thaliana}, partial (16%)
Length = 762
Score = 45.8 bits (107), Expect = 5e-05
Identities = 23/39 (58%), Positives = 25/39 (63%)
Frame = -3
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRG 641
GG GGRG GGG G ARG G+G+G G GRG GRG
Sbjct: 436 GGGGGRGRVSGGGRGGARGRGQGQGWGRGRGSGRGRGRG 320
Score = 40.4 bits (93), Expect = 0.002
Identities = 23/43 (53%), Positives = 23/43 (53%), Gaps = 6/43 (13%)
Frame = -3
Query: 609 GPWRGGGGGR------ARGGYYGRGYGGYGGRGRGGGRGMPGG 645
G GGGGGR RGG GRG G GRGRG GRG G
Sbjct: 448 GGLSGGGGGRGRVSGGGRGGARGRGQGQGWGRGRGSGRGRGRG 320
Score = 30.8 bits (68), Expect = 1.7
Identities = 15/27 (55%), Positives = 16/27 (58%)
Frame = -3
Query: 602 RGGWGGRGPWRGGGGGRARGGYYGRGY 628
RGG GRG +G G GR G GRGY
Sbjct: 397 RGGARGRGQGQGWGRGRGSGRGRGRGY 317
Score = 30.4 bits (67), Expect = 2.2
Identities = 13/37 (35%), Positives = 17/37 (45%)
Frame = +3
Query: 212 QQPMPSSLQQPLPSSMQQPMPSSMQQPMPSSMQQPMP 248
Q P+P L PLP P P + P+P P+P
Sbjct: 315 QYPLPLPLPLPLPRPQPWPWPRPLAPPLPPPETLPLP 425
Score = 29.6 bits (65), Expect = 3.8
Identities = 19/54 (35%), Positives = 23/54 (42%)
Frame = +3
Query: 190 LPQLHQQSLFRPPPGLSMPSSMQQPMPSSLQQPLPSSMQQPMPSSMQQPMPSSM 243
L Q HQ L P P L +P P P L PLP P+P PS++
Sbjct: 300 LKQNHQYPLPLPLP-LPLPRPQPWPWPRPLAPPLPPPETLPLPPPPPDNPPSAL 458
>TC87476 homologue to PIR|T11622|T11622 extensin class 1 precursor - cowpea,
partial (48%)
Length = 939
Score = 45.4 bits (106), Expect = 7e-05
Identities = 38/126 (30%), Positives = 47/126 (37%)
Frame = +3
Query: 208 PSSMQQPMPSSLQQPLPSSMQQPMPSSMQQPMPSSMQQPMPSSMQQPMLSSMQQPLQFPS 267
P S P P + P P S P P + P P S P P + P S P P
Sbjct: 369 PPSPSPPPPYVYKSPPPPSPSPPPPYVYKSPPPPSPSPPPPYVYKSPPPPSASPP---PP 539
Query: 268 FSSSLPTGPSNFPEFPSAFLPVGTSSPNITSTSAPPLNLSTTILPPAPSATLAPETFQAS 327
+ P PS PS P G SP S S PP + + PP+PS P +
Sbjct: 540 YYYKSPPPPS-----PSPPPPYGYKSPPPPSPSPPPPYIYKSPPPPSPS----PPPYHPY 692
Query: 328 VSNKAP 333
+ N P
Sbjct: 693 LYNSPP 710
Score = 42.4 bits (98), Expect = 6e-04
Identities = 34/119 (28%), Positives = 46/119 (38%)
Frame = +2
Query: 198 LFRPPPGLSMPSSMQQPMPSSLQQPLPSSMQQPMPSSMQQPMPSSMQQPMPSSMQQPMLS 257
+++ PP P S P P + P P S P P + P P S P P + P
Sbjct: 14 IYKSPP----PPSPSPPPPYVYKSPPPPSPSPPPPYVYKSPPPPSPSPPPPYVYKSPPPP 181
Query: 258 SMQQPLQFPSFSSSLPTGPSNFPEFPSAFLPVGTSSPNITSTSAPPLNLSTTILPPAPS 316
+ P P + P PS P P + SP S S PP + + PP+PS
Sbjct: 182 TPSPP---PPYIYKSPPPPSPSPPPPYVY-----KSPPPPSPSPPPPYVYKSPPPPSPS 334
Score = 35.0 bits (79), Expect = 0.090
Identities = 55/198 (27%), Positives = 84/198 (41%), Gaps = 9/198 (4%)
Frame = +2
Query: 201 PPPGLSMP-----SSMQQPMPSSLQQPLPSSMQQPMPSSMQQPMPSSMQQPMPSSMQQP- 254
PPP S P S P PS P P + P P S P P + P P S P
Sbjct: 173 PPPTPSPPPPYIYKSPPPPSPS---PPPPYVYKSPPPPSPSPPPPYVYKSPPPPSPSPPP 343
Query: 255 --MLSSMQQPLQFPSFSSSLPTGPSNFPEFPSAFLPVGTSSPNITSTSAPPLNLSTTILP 312
+ S + F + S L S+F PS+ + + SSP I+ T P L +
Sbjct: 344 PYIYKSPPTTIPFTTTSICL*VPSSSFSFTPSS-ICLQISSPTISFT---PSTLCL*VTS 511
Query: 313 PAPSATLAPETFQASVSNKAPTVSLPAATLGANLPSLA-PFTSGGSDINAAVPLANKPNA 371
+ S T + Q S S T S+ ++T+ + + + FT+ A++ L +
Sbjct: 512 TSFSFTTSSILLQISTS----TFSITSSTIWL*VTATSFAFTT------ASLYLQVSTST 661
Query: 372 ISGSSLSYQTVPQQMTPS 389
IS +S S ++P Q+TP+
Sbjct: 662 ISITS-SISSLPLQLTPA 712
Score = 30.4 bits (67), Expect = 2.2
Identities = 19/43 (44%), Positives = 20/43 (46%), Gaps = 6/43 (13%)
Frame = -3
Query: 605 WGGRGPWRGGGGGRARGGYYGRGYGG------YGGRGRGGGRG 641
+GG G GGGG G G G GG YGG G G G G
Sbjct: 541 YGGGGEAEGGGGDL*T*GGGGEGDGGGGDL*TYGGGGEGEGGG 413
Score = 30.0 bits (66), Expect = 2.9
Identities = 19/43 (44%), Positives = 20/43 (46%), Gaps = 6/43 (13%)
Frame = -1
Query: 605 WGGRGPWRGGGGGRARGGYYGRGYGG------YGGRGRGGGRG 641
+GG G GGGG G G G GG YGG G G G G
Sbjct: 348 YGGGGEGDGGGGDL*TYGGGGDGDGGGGDL*TYGGGGEGDGGG 220
Score = 29.6 bits (65), Expect = 3.8
Identities = 26/82 (31%), Positives = 32/82 (38%), Gaps = 14/82 (17%)
Frame = -1
Query: 578 QNGRVRYSEQIKMDTETFGDFSRHRGG------------WGGRGPWRGGGGGRARGGYYG 625
+ G R+ E + GD + GG +GG G GGGG G G
Sbjct: 417 EEGTYRHMEVVVKGMVVGGDL*IYGGGGEGDGGGGDL*TYGGGGDGDGGGGDL*TYGGGG 238
Query: 626 RGYGGYGGR--GRGGGRGMPGG 645
G GG G GGG G+ GG
Sbjct: 237 EGDGGGGDL*I*GGGGEGVGGG 172
Score = 29.6 bits (65), Expect = 3.8
Identities = 19/43 (44%), Positives = 20/43 (46%), Gaps = 6/43 (13%)
Frame = -1
Query: 605 WGGRGPWRGGGGGRARGGYYGRGYGG------YGGRGRGGGRG 641
+GG G GGGG G G G GG YGG G G G G
Sbjct: 156 YGGGGDGEGGGGLL*TYGGGGDGDGGGGDLYTYGGGGEGDGGG 28
Score = 29.3 bits (64), Expect = 4.9
Identities = 21/49 (42%), Positives = 23/49 (46%)
Frame = -1
Query: 591 DTETFGDFSRHRGGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGG 639
D T+G GG G + GGG G GG YGG GG G GGG
Sbjct: 168 DL*TYGGGGDGEGGGGLL*TYGGGGDGDGGGGDL-YTYGG-GGEGDGGG 28
Score = 29.3 bits (64), Expect = 4.9
Identities = 19/43 (44%), Positives = 20/43 (46%), Gaps = 6/43 (13%)
Frame = -1
Query: 605 WGGRGPWRGGGGGRARGGYYGRGYGG------YGGRGRGGGRG 641
+GG G GGGG G G G GG YGG G G G G
Sbjct: 252 YGGGGEGDGGGGDL*I*GGGGEGVGGGGDL*TYGGGGDGEGGG 124
>TC84888 similar to GP|21322752|dbj|BAB78536. cold shock protein-1 {Triticum
aestivum}, partial (58%)
Length = 612
Score = 45.4 bits (106), Expect = 7e-05
Identities = 28/57 (49%), Positives = 33/57 (57%), Gaps = 5/57 (8%)
Frame = +2
Query: 596 GDFSRHRGGWGGR--GPWRGG-GGGRARGGYYGRGYGGYGGRGRG--GGRGMPGGRY 647
G + + GG GG G + GG GGGR GGY GYGG GG +G GG+G GG Y
Sbjct: 341 GGYDQGYGGGGGSYGGGFGGGRGGGRGGGGYXQGGYGGGGGYXQGGYGGQGGYGGGY 511
Score = 44.7 bits (104), Expect = 1e-04
Identities = 26/45 (57%), Positives = 27/45 (59%), Gaps = 10/45 (22%)
Frame = +2
Query: 613 GGGGGRARGGY---YGRGYGGYGGR-------GRGGGRGMPGGRY 647
GGGGGR GGY Y +GYGG GG GRGGGRG GG Y
Sbjct: 305 GGGGGRGGGGYGGGYDQGYGGGGGSYGGGFGGGRGGGRG--GGGY 433
Score = 40.0 bits (92), Expect = 0.003
Identities = 25/58 (43%), Positives = 28/58 (48%), Gaps = 6/58 (10%)
Frame = +2
Query: 596 GDFSRHRGGWGGRGPWRGGGGGRARGGYY------GRGYGGYGGRGRGGGRGMPGGRY 647
G R GG+GG GGGG + GG + GRG GGY G GGG G G Y
Sbjct: 308 GGGGRGGGGYGGGYDQGYGGGGGSYGGGFGGGRGGGRGGGGYXQGGYGGGGGYXQGGY 481
>BG648523 similar to GP|9955540|emb| putative protein {Arabidopsis thaliana},
partial (9%)
Length = 770
Score = 45.1 bits (105), Expect = 9e-05
Identities = 45/153 (29%), Positives = 58/153 (37%), Gaps = 11/153 (7%)
Frame = +3
Query: 153 PGGNVVSWGASSPAP-----------NANGGGLAMPMYWQGYYGAPNGLPQLHQQSLFRP 201
PG V+ G ++P P G G+AMP+ + G+ Q H S+ P
Sbjct: 231 PGHFVMPEGPTTPGPFPPGLMTRNEGTIPGVGVAMPLSIPSFDGSQGEQKQPHPGSIGAP 410
Query: 202 PPGLSMPSSMQQPMPSSLQQPLPSSMQQPMPSSMQQPMPSSMQQPMPSSMQQPMLSSMQQ 261
P S+ P + QQP + QQ +P QQ PMP +M Q SS
Sbjct: 411 PLPPGPHPSLLNP---NQQQPFQQNPQQ-IPQHQQQLQQHMGPLPMPPNMPQIQHSSHSS 578
Query: 262 PLQFPSFSSSLPTGPSNFPEFPSAFLPVGTSSP 294
L LP P P LPV TS P
Sbjct: 579 MLP----HQHLPRPPPQMPHGMPGSLPVPTSHP 665
Score = 31.6 bits (70), Expect = 0.99
Identities = 21/51 (41%), Positives = 27/51 (52%), Gaps = 3/51 (5%)
Frame = +3
Query: 193 LHQQSLFRPPPGL--SMPSSMQQPMPSSLQQPLPSSM-QQPMPSSMQQPMP 240
L Q L RPPP + MP S+ P+P+S P+P M Q + M PMP
Sbjct: 582 LPHQHLPRPPPQMPHGMPGSL--PVPTSHPMPIPGPMGMQGTMNQMGPPMP 728
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.310 0.130 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,456,658
Number of Sequences: 36976
Number of extensions: 353851
Number of successful extensions: 7725
Number of sequences better than 10.0: 530
Number of HSP's better than 10.0 without gapping: 3774
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6007
length of query: 647
length of database: 9,014,727
effective HSP length: 102
effective length of query: 545
effective length of database: 5,243,175
effective search space: 2857530375
effective search space used: 2857530375
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146570.8


