

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146570.7 - phase: 0
(915 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
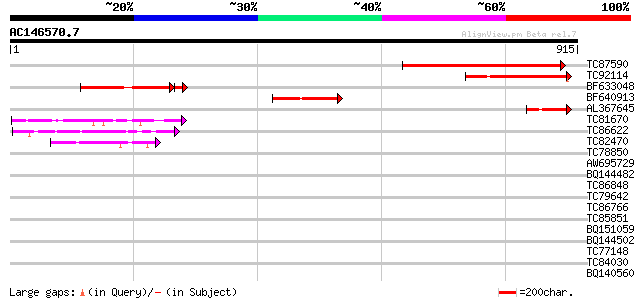
Score E
Sequences producing significant alignments: (bits) Value
TC87590 weakly similar to PIR|G96522|G96522 F11A17.16 [imported]... 238 8e-63
TC92114 similar to GP|11994760|dbj|BAB03089. contains similarity... 218 8e-57
BF633048 similar to GP|11994760|d contains similarity to pheroph... 184 2e-53
BF640913 similar to GP|11994760|db contains similarity to pherop... 131 1e-30
AL367645 similar to GP|22135814|gb| AT4g18560/F28J12_220 {Arabid... 77 3e-14
TC81670 weakly similar to PIR|E86293|E86293 hypothetical protein... 52 8e-07
TC86622 weakly similar to GP|20197623|gb|AAM15156.1 putative myo... 46 6e-05
TC82470 homologue to GP|23497460|gb|AAN37003.1 hypothetical prot... 45 1e-04
TC78850 homologue to GP|10177413|dbj|BAB10544. fibrillarin-like ... 41 0.002
AW695729 similar to GP|22135814|gb| AT4g18560/F28J12_220 {Arabid... 40 0.003
BQ144482 similar to GP|6523547|emb| hydroxyproline-rich glycopro... 40 0.004
TC86848 similar to GP|10177413|dbj|BAB10544. fibrillarin-like {A... 40 0.005
TC79642 similar to PIR|T07397|T07397 kinesin heavy chain-like pr... 40 0.005
TC86766 weakly similar to PIR|T06291|T06291 extensin homolog T9E... 39 0.009
TC85851 similar to PIR|T12180|T12180 probable transcription fact... 39 0.009
BQ151059 similar to GP|7291525|gb|A CG9888-PA {Drosophila melano... 39 0.012
BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich gl... 38 0.015
TC77148 ENOD20 38 0.015
TC84030 38 0.015
BQ140560 weakly similar to GP|15128223|db hypothetical protein~s... 38 0.015
>TC87590 weakly similar to PIR|G96522|G96522 F11A17.16 [imported] -
Arabidopsis thaliana, partial (45%)
Length = 2425
Score = 238 bits (607), Expect = 8e-63
Identities = 115/264 (43%), Positives = 179/264 (67%), Gaps = 1/264 (0%)
Frame = +1
Query: 634 VKRAPELVEFYQSLMKREAKKDASLLTSSTSNAADTRSN-VIAEIENRSSFLLAVKADVE 692
+KR +L ++ R KKD + ++ N ++ EI+NRS+ LLA++ D++
Sbjct: 1147 LKRLLQLSNYFTP*RIRTPKKDLKGSINHQKPITNSAHNSIVGEIQNRSAHLLAIREDIQ 1326
Query: 693 TQGDFVMSLATEVRAASFSKIEDVVAFVNWLDEELSFLSDERAVLKHFDWPEGKSDALRE 752
T+G+F+ L +V AS+ IEDV+ FV+WLD ELS L+DERAVLKHF WPE K+D +RE
Sbjct: 1327 TKGEFINGLINKVVDASYVDIEDVLKFVDWLDGELSTLADERAVLKHFKWPERKADTMRE 1506
Query: 753 ASFEYQDLMKLEKQVSNFTDDPKLPCEDALQKMYSLLEKLEQSVYALLRTRDFAISRYKE 812
A+ EY++L LE+++S++ DDP +PC +L+K+ SLL+K E+S+ L+ R+ I Y+
Sbjct: 1507 AAVEYRELKMLEQEISSYKDDPDIPCVASLKKIASLLDKSERSIQKLIVLRNSVIRSYQM 1686
Query: 813 FGVPVNWLLDSGVVGKIKLSSVQLANKYMKRIASEIDTLSGPENEPTREFLILQGVRFSF 872
+ +P W+LDSG+ KIK SS+ L YMKR+ E++++ + E ++ L+LQGV F++
Sbjct: 1687 YNIPTAWMLDSGISSKIKQSSMTLVKMYMKRLTMELESIRNSDRESNQDSLLLQGVHFAY 1866
Query: 873 RVHQFAGGFDTESMKAFEELRNNI 896
R HQFAGG D+E++ AFEE+R +
Sbjct: 1867 RAHQFAGGLDSETLCAFEEIRQRV 1938
Score = 31.6 bits (70), Expect = 1.4
Identities = 22/106 (20%), Positives = 46/106 (42%), Gaps = 10/106 (9%)
Frame = +3
Query: 559 NKQSEDVKDFDNQTITEMKLFKIEKRP--------PRLPRPPPKPSDGAPVSNSLNEIP- 609
N + ++K + +T K + +K+P +P P P + +++ + P
Sbjct: 891 NARVLNLKTYKKSLLTNWKCLR*KKKPILRVIFVKSSIPAPIPNHAAIREITSLGRKSPP 1070
Query: 610 -YAPSVPSPPPPPGSLPRGAVGDDKVKRAPELVEFYQSLMKREAKK 654
+ P PPPPP R ++AP +V+ + SL ++ ++
Sbjct: 1071NHCLMPPPPPPPPPIPSRPLAKLANTQKAPAVVQLFHSLKNQDTEE 1208
>TC92114 similar to GP|11994760|dbj|BAB03089. contains similarity to
pherophorin~gene_id:T5M7.14 {Arabidopsis thaliana},
partial (15%)
Length = 674
Score = 218 bits (555), Expect = 8e-57
Identities = 115/178 (64%), Positives = 137/178 (76%), Gaps = 7/178 (3%)
Frame = +3
Query: 736 VLKHFDWPEGKSDALREASFEYQDL---MKLEKQVSNFTDDPKLPCEDALQKMYSLLEKL 792
VLKHFDWPEGK+DA + S + ++E S+ P + L+KMYSLLEK+
Sbjct: 3 VLKHFDWPEGKADAPKGGSLLNIKI**NWRIESLPSSMI--PNSHVKLRLKKMYSLLEKV 176
Query: 793 EQSVYALLRTRDFAISRYKEFGVPVNWLLDSGVVGKIKLSSVQLANKYMKRIASEIDTLS 852
EQSVYALLRT+D AISRY+EFG+P+NWL D+GVVGKIKLSSVQLA KYMKR+ASE+D LS
Sbjct: 177 EQSVYALLRTKDMAISRYREFGIPINWLQDAGVVGKIKLSSVQLARKYMKRVASELDALS 356
Query: 853 GPENEPTREFLILQGVRFSFRVHQFAGGFDTESMKAFEELRNNIHV----QAGEYNNK 906
GPE EP REFLILQGVRF+FRVHQFAGGFD ESMKAFE+LR+ I Q G+ ++K
Sbjct: 357 GPEKEPAREFLILQGVRFAFRVHQFAGGFDAESMKAFEDLRSRIQTPQAPQVGDEDSK 530
>BF633048 similar to GP|11994760|d contains similarity to
pherophorin~gene_id:T5M7.14 {Arabidopsis thaliana},
partial (15%)
Length = 510
Score = 184 bits (468), Expect(2) = 2e-53
Identities = 102/151 (67%), Positives = 117/151 (76%)
Frame = +2
Query: 115 SLQEENNKLQEELIHEASAKRELEVARNKIKELQRQIKIIANQTKGQLLLLKQKVSGLQA 174
SLQ E KLQEEL + ASAKR+LE+ARNKIKELQRQ+++ ANQTKGQLLLLKQ+VSGLQ
Sbjct: 5 SLQAERKKLQEELTNGASAKRDLELARNKIKELQRQMQLEANQTKGQLLLLKQQVSGLQV 184
Query: 175 KEEEVVKKDAEIENNLKTVNDFEIEKKLKTVNDLEIEAVGLKRRNKELQHEKRELTVKLN 234
KEE KDA EI+KKLK VNDLE+ V LKR+NKELQ+EKRELTVKLN
Sbjct: 185 KEEAGAIKDA------------EIDKKLKAVNDLEVAVVELKRKNKELQYEKRELTVKLN 328
Query: 235 AAESRITELSSVTENEMIADAKSETGRLRHA 265
AAESR+ ELS++TE EM+A AK E LRHA
Sbjct: 329 AAESRVAELSNMTETEMVAKAKEEVSNLRHA 421
Score = 44.3 bits (103), Expect(2) = 2e-53
Identities = 20/22 (90%), Positives = 21/22 (94%)
Frame = +3
Query: 266 NEDLQKQVEGLQMNRFSEVEEL 287
NEDL KQVEGLQMNRFSE+EEL
Sbjct: 423 NEDLSKQVEGLQMNRFSEIEEL 488
Score = 39.7 bits (91), Expect = 0.005
Identities = 40/153 (26%), Positives = 76/153 (49%), Gaps = 12/153 (7%)
Frame = +2
Query: 48 EMAKNGSDS--EIEWLRNVVEELEEREMKLQSELL--EYYSLKEQVPVIEEFQRQLRIKS 103
E NG+ + ++E RN ++EL+ R+M+L++ + LK+QV ++ + IK
Sbjct: 35 EELTNGASAKRDLELARNKIKELQ-RQMQLEANQTKGQLLLLKQQVSGLQVKEEAGAIKD 211
Query: 104 VEIDM-------LHMTIKSLQEENNKLQEELIHEASAKRELEVARNKIKELQRQIKIIAN 156
EID L + + L+ +N +LQ E KREL V K+ + ++ ++N
Sbjct: 212 AEIDKKLKAVNDLEVAVVELKRKNKELQYE-------KRELTV---KLNAAESRVAELSN 361
Query: 157 QTKGQLLL-LKQKVSGLQAKEEEVVKKDAEIEN 188
T+ +++ K++VS L+ + V+ N
Sbjct: 362 MTETEMVAKAKEEVSNLRHAK*RPVEASGRTSN 460
>BF640913 similar to GP|11994760|db contains similarity to
pherophorin~gene_id:T5M7.14 {Arabidopsis thaliana},
partial (7%)
Length = 424
Score = 131 bits (329), Expect = 1e-30
Identities = 75/116 (64%), Positives = 90/116 (76%), Gaps = 3/116 (2%)
Frame = +1
Query: 425 LESLMLRNAHDAVAITTFEQKEHKPTHSPETPFLPS--LRKVSSSDDILDSVSASFQLMS 482
LESLM+RNA D+VAITTF Q + + +SPETP S LR+V+SSD L+SV++SF LMS
Sbjct: 46 LESLMIRNASDSVAITTFGQGDQESIYSPETPNTASAGLRRVTSSDS-LNSVASSFHLMS 222
Query: 483 KS-VDRSLDEKYPAYKDLHKLALAREKQLKEKAEKARGEKFGDNSNLNTTKAEEER 537
KS VD S+DEKYPAYKD HKLA+ARE LKEKAEKAR FG++S+LN T E ER
Sbjct: 223 KSSVDASVDEKYPAYKDRHKLAMARESDLKEKAEKARVXNFGNSSSLNMTXIERER 390
>AL367645 similar to GP|22135814|gb| AT4g18560/F28J12_220 {Arabidopsis
thaliana}, partial (10%)
Length = 522
Score = 77.0 bits (188), Expect = 3e-14
Identities = 39/75 (52%), Positives = 55/75 (73%), Gaps = 3/75 (4%)
Frame = +1
Query: 835 QLANKYMKRIASEIDTLSGPENEPTREFLILQGVRFSFRVHQFAGGFDTESMKAFEELRN 894
+LA KYMKR+++E++T+ G P E LI+QGVRF+FRVHQFA GFD ++M+AF+ELR+
Sbjct: 1 KLAMKYMKRVSAELETVGGG---PEEEELIVQGVRFAFRVHQFASGFDADTMRAFQELRD 171
Query: 895 ---NIHVQAGEYNNK 906
+ HVQ + K
Sbjct: 172 KARSCHVQCHDQQQK 216
>TC81670 weakly similar to PIR|E86293|E86293 hypothetical protein AAF18488.1
[imported] - Arabidopsis thaliana, partial (41%)
Length = 1797
Score = 52.4 bits (124), Expect = 8e-07
Identities = 73/309 (23%), Positives = 135/309 (43%), Gaps = 27/309 (8%)
Frame = +2
Query: 4 IMNLANDFD--DEILSEFESLLSEQTDFPLLSEKTDDSKKHGGNETEMAKNGSDSEIEWL 61
+ NLAN+ D +E L++ + +E+T LS ++ + E +++ E +
Sbjct: 821 VENLANEIDITNENLNKMQYKYNEKT--MSLSRMLEEKDRLHNAFVEESRSMQRKAREEV 994
Query: 62 RNVVEELEEREMKLQSELLEYYSLKEQVPVIEEFQRQLRIKSVEIDMLHMTIKSLQEENN 121
R ++EE E KL++EL E++ ++ + R L + V D ++ +++ +
Sbjct: 995 RRILEEQE----KLRNEL------DEKMRKLDTWSRDLNKREVLTDQERQKLEEDKKKKD 1144
Query: 122 KLQEELIHEASA---------------KRELEVARNKIKELQRQ------IKIIANQTKG 160
E L+ + KRE E A NKI +L++Q +++ + +G
Sbjct: 1145 SRNESLMLASKEQKIADENVFRLVEEQKREKEEALNKILQLEKQLDAKQKLEMEIEELRG 1324
Query: 161 QLLLLKQKVSGLQAKEEEVVKKDAEIENNLKTVNDFEIEKKLKTVNDLE----IEAVGLK 216
+L ++K L +++ +KK E N+ E+E K++++ D+E V +
Sbjct: 1325 KLQVMKH----LGDQDDTAIKKKMEEMNS-------ELEDKIESLEDMESMNSTLIVKER 1471
Query: 217 RRNKELQHEKRELTVKLNAAESRITELSSVTENEMIADAKSETGRLRHANEDLQKQVEGL 276
+ N ELQ ++EL L NEM+ AK+ G R + D QK
Sbjct: 1472 QSNDELQEARKELIEGL---------------NEMLTGAKTNIGTKRMGDLD-QKVFVNA 1603
Query: 277 QMNRFSEVE 285
RFS E
Sbjct: 1604 CKKRFSSDE 1630
>TC86622 weakly similar to GP|20197623|gb|AAM15156.1 putative myosin heavy
chain {Arabidopsis thaliana}, partial (17%)
Length = 2546
Score = 46.2 bits (108), Expect = 6e-05
Identities = 62/281 (22%), Positives = 125/281 (44%), Gaps = 11/281 (3%)
Frame = +2
Query: 5 MNLANDFDDEILSEFESLLSEQTDFP----LLSEKTDDSKKHGGNETEMAKNGSDSEIEW 60
+ AN+ + E+ ++ E+ LSEK +S+ N E+ ++ +
Sbjct: 128 LQTANESEIELKESLNAVTDEKKKLEDALNSLSEKLAESE----NLLEIVRDDLNLTQVK 295
Query: 61 LRNVVEELEEREMKLQSELLEYYSLKEQVPVIEEFQRQLR-IKSVEIDMLHMTIKSLQEE 119
L++ +L+ E++ +SE+ E ++ E+ + +L +++E++ LH ++ ++
Sbjct: 296 LQSTENDLKAAELR-ESEIREKHNAIEENLAVRGRDIELTSARNLELESLHESLT--RDS 466
Query: 120 NNKLQEELIHEASAKRELEVARNKIKELQRQIKIIANQT---KGQLLLLKQKVSGLQAKE 176
KLQE + S E++ KIK L+ I Q+ K + K++ LQ++
Sbjct: 467 EQKLQEAIEKFNSKDSEVQSLLEKIKILEENIAGAGEQSISLKSEFEESLSKLASLQSEN 646
Query: 177 EEVVKKDAEIENNLKTVNDFEIEKKLKTVNDLEIEAVGLKRRNKELQHEKRELTVKLNAA 236
E++ ++ E E KT F + L N + LK + ELQ + +
Sbjct: 647 EDLKRQIVEAEK--KTSQSFSENELLVGTN------IQLKTKIDELQESLNSV-----VS 787
Query: 237 ESRITELSSVTENEMIA---DAKSETGRLRHANEDLQKQVE 274
E +T V+ ++A D +S++ + ANE +VE
Sbjct: 788 EKEVTAQELVSHKNLLAELNDVQSKSSEIHSANEVRILEVE 910
Score = 38.9 bits (89), Expect = 0.009
Identities = 56/213 (26%), Positives = 89/213 (41%), Gaps = 14/213 (6%)
Frame = +2
Query: 5 MNLANDFDDEILSEFESLLSEQTDFPLLSEKTDDSKKHGGNETEMAKNGSDSEIEWLRNV 64
++L ++F+ E LS+ SL SE D + + +E E+ G++ + L+
Sbjct: 584 ISLKSEFE-ESLSKLASLQSENEDLKRQIVEAEKKTSQSFSENELLV-GTNIQ---LKTK 748
Query: 65 VEELEEREMKLQSE----LLEYYSLKEQVPVIEEFQRQLRIKSVEIDMLHMTIKSLQEEN 120
++EL+E + SE E S K + + + Q KS EI + + E
Sbjct: 749 IDELQESLNSVVSEKEVTAQELVSHKNLLAELNDVQS----KSSEIHSANEV--RILEVE 910
Query: 121 NKLQEELIHEASAKRELEVARNKIKELQRQIKIIANQT----------KGQLLLLKQKVS 170
+KLQE L + E + K+ L+ QIKI Q K +L K+
Sbjct: 911 SKLQEALQKHTEKESETKELNEKLNTLEGQIKIYEEQAHEAVAAAENRKAELEESLIKLK 1090
Query: 171 GLQAKEEEVVKKDAEIENNLKTVNDFEIEKKLK 203
L+A EE K E E +N E+KLK
Sbjct: 1091HLEAAVEEQQNKSLERETETAGIN----EEKLK 1177
Score = 36.6 bits (83), Expect = 0.045
Identities = 96/430 (22%), Positives = 163/430 (37%), Gaps = 20/430 (4%)
Frame = +2
Query: 169 VSGLQAKEEEVVKKDAEIENNLKTVNDFEIEKKLKTVNDLEIEAVGLKRRNKELQHEKRE 228
VS L ++ E + + +EN L+T N+ EIE LK + EK++
Sbjct: 65 VSDLTSELESFKVRTSSLENTLQTANESEIE---------------LKESLNAVTDEKKK 199
Query: 229 LTVKLNAAESRITELSSVTENEMIADAKSETG-RLRHANEDLQ--KQVEGLQMNRFSEVE 285
L LN+ ++ E ++ E++ D + T +L+ DL+ + E + + +E
Sbjct: 200 LEDALNSLSEKLAESENLL--EIVRDDLNLTQVKLQSTENDLKAAELRESEIREKHNAIE 373
Query: 286 ELVYLRWVNACLRYELRNYKAPS-GKSLARDLN-------NSFNPKSQ------EKAKQL 331
E + +R + L RN + S +SL RD FN K EK K L
Sbjct: 374 ENLAVRGRDIELT-SARNLELESLHESLTRDSEQKLQEAIEKFNSKDSEVQSLLEKIKIL 550
Query: 332 MLEYAGSERGHGDTDIESNFSHDHSSPGSEDLDNAYINSPTYKYSNLSKKTSLIQKLKKW 391
AG+ G ++S F S S +N + K +++ KK
Sbjct: 551 EENIAGA--GEQSISLKSEFEESLSKLASLQSENEDL------------KRQIVEAEKK- 685
Query: 392 NKNNDSSAFSSPARSLSIGSPCRVSTSY-RPRNTLESLMLRNAHDAVAITTFEQKEHKPT 450
S +FS L +G+ ++ T + +L S++ +T E HK
Sbjct: 686 ----TSQSFSE--NELLVGTNIQLKTKIDELQESLNSVVSEK-----EVTAQELVSHK-- 826
Query: 451 HSPETPFLPSLRKVSSSDDILDSVSASFQLMSKSVDRSLDEKYPAYKDLHKLALAREKQL 510
L L V S + S + ++ V+ L E + + + K+L
Sbjct: 827 -----NLLAELNDVQSKSSEIHSAN---EVRILEVESKLQEALQKHTEKE----SETKEL 970
Query: 511 KEKAEKARGE-KFGDNSNLNTTKAEEERSTSLDSELTQVNE-KACVSDGLNKQSEDVKDF 568
EK G+ K + A E R L+ L ++ +A V + NK E ++
Sbjct: 971 NEKLNTLEGQIKIYEEQAHEAVAAAENRKAELEESLIKLKHLEAAVEEQQNKSLE--RET 1144
Query: 569 DNQTITEMKL 578
+ I E KL
Sbjct: 1145ETAGINEEKL 1174
>TC82470 homologue to GP|23497460|gb|AAN37003.1 hypothetical protein
{Plasmodium falciparum 3D7}, partial (8%)
Length = 1064
Score = 45.1 bits (105), Expect = 1e-04
Identities = 43/189 (22%), Positives = 83/189 (43%), Gaps = 11/189 (5%)
Frame = +2
Query: 66 EELEEREMKLQSELLEYYSLKEQVPVIEEFQRQLRIKSVEIDMLHMTIKSLQEENNKLQE 125
+EL+ +E++L+ + KE ++E L K + IK L+ + +L+
Sbjct: 386 KELKTKELELRQVMDNISKQKELESQVKELVNDLVSKQKHFES---HIKELESKERQLEG 556
Query: 126 ELIHEASAKRELEVARNKIKELQRQIKIIANQTKGQLLLLKQKVSGLQAKEE-------E 178
L ++E E N+++ +R K + +L+ LK ++ L +KE+ E
Sbjct: 557 RLKEHELEEKEFEGRMNELESKERHFKSEVEEINAKLMPLKGQIKELASKEKQLNGQVKE 736
Query: 179 VVKKDAEIENNLKTVNDFEIEKKLKTVNDLEIEAVGLKRRNK----ELQHEKRELTVKLN 234
+ K + EN +K E+E K K E +R + E Q +K+ +++
Sbjct: 737 LESKKNQFENRIK-----ELESKEKQHEGRVKEHASKEREFESQVMEQQFKKKLFEIQVK 901
Query: 235 AAESRITEL 243
A ES+ +L
Sbjct: 902 ALESKENQL 928
>TC78850 homologue to GP|10177413|dbj|BAB10544. fibrillarin-like
{Arabidopsis thaliana}, partial (87%)
Length = 1177
Score = 41.2 bits (95), Expect = 0.002
Identities = 19/37 (51%), Positives = 19/37 (51%)
Frame = -1
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPP 621
PPR P PPPKP P L P P P PPPPP
Sbjct: 208 PPRAPLPPPKP---LPPPRGLKSPPLPPPKPLPPPPP 107
>AW695729 similar to GP|22135814|gb| AT4g18560/F28J12_220 {Arabidopsis
thaliana}, partial (7%)
Length = 408
Score = 40.4 bits (93), Expect = 0.003
Identities = 19/44 (43%), Positives = 28/44 (63%)
Frame = +3
Query: 610 YAPSVPSPPPPPGSLPRGAVGDDKVKRAPELVEFYQSLMKREAK 653
+A S P PP G +P KV++ PE+VEFY SLM+R+++
Sbjct: 252 HAHSTPPPPAKGGKMPPA-----KVRKVPEVVEFYHSLMRRDSQ 368
>BQ144482 similar to GP|6523547|emb| hydroxyproline-rich glycoprotein DZ-HRGP
{Volvox carteri f. nagariensis}, partial (16%)
Length = 993
Score = 40.0 bits (92), Expect = 0.004
Identities = 20/39 (51%), Positives = 22/39 (56%)
Frame = +3
Query: 583 KRPPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPP 621
K PR P PPP P P S S + P +P PSPPPPP
Sbjct: 168 KVAPRPPFPPPLPFYLRPPSPSPSPPPLSPPSPSPPPPP 284
Score = 38.9 bits (89), Expect = 0.009
Identities = 23/48 (47%), Positives = 24/48 (49%), Gaps = 7/48 (14%)
Frame = +1
Query: 581 IEKRPPRLP-----RPPPKPSDGAPVSNSLNEI--PYAPSVPSPPPPP 621
I PP LP RPPP P P+S SL P AP P PPPPP
Sbjct: 211 ISAPPPPLPPLPLFRPPPPPPPPPPLSLSLAPAFAPPAPGPPRPPPPP 354
Score = 38.9 bits (89), Expect = 0.009
Identities = 23/46 (50%), Positives = 26/46 (56%), Gaps = 5/46 (10%)
Frame = +1
Query: 584 RPPRLPR----PPPKPSDGAPVSNSLNEIP-YAPSVPSPPPPPGSL 624
RPP+ PR PPP PS AP L +P + P P PPPPP SL
Sbjct: 160 RPPKSPRAHLFPPPSPSISAPPP-PLPPLPLFRPPPPPPPPPPLSL 294
Score = 33.9 bits (76), Expect = 0.29
Identities = 17/40 (42%), Positives = 19/40 (47%), Gaps = 2/40 (5%)
Frame = +2
Query: 588 LPRPPPKPSDGAPVSNSLNEIPYAPSV--PSPPPPPGSLP 625
LP PPP P +P P AP P P PPPG +P
Sbjct: 260 LPLPPPPPPSASPWPRPSPPRPPAPRAPPPRPGPPPGPVP 379
Score = 31.6 bits (70), Expect = 1.4
Identities = 16/44 (36%), Positives = 20/44 (45%)
Frame = +2
Query: 582 EKRPPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
+ RP PPP P+ SL +AP PPPPP + P
Sbjct: 167 QSRPAPTFSPPPPLLSPPPLPLSLPSPSFAPLPLPPPPPPSASP 298
>TC86848 similar to GP|10177413|dbj|BAB10544. fibrillarin-like {Arabidopsis
thaliana}, partial (92%)
Length = 1250
Score = 39.7 bits (91), Expect = 0.005
Identities = 18/41 (43%), Positives = 22/41 (52%)
Frame = -2
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PPR P PP P P +LN +P +P +P PP P LP
Sbjct: 244 PPRPPPRPPLPPPPLPPPLALNGVPLSPPLPPKPPLPPPLP 122
Score = 30.0 bits (66), Expect = 4.2
Identities = 18/41 (43%), Positives = 20/41 (47%), Gaps = 1/41 (2%)
Frame = -2
Query: 586 PRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPP-PPGSLP 625
P P PPP +G P+S P P P PPP PP S P
Sbjct: 214 PPPPLPPPLALNGVPLSP-----PLPPKPPLPPPLPPLSPP 107
>TC79642 similar to PIR|T07397|T07397 kinesin heavy chain-like protein
(clone PKCBP) - potato, partial (42%)
Length = 2321
Score = 39.7 bits (91), Expect = 0.005
Identities = 41/174 (23%), Positives = 91/174 (51%), Gaps = 12/174 (6%)
Frame = +2
Query: 56 SEIEWLRNVVEELEEREMKLQSELLEYYSLKEQVP----VIEEFQRQLRIKSVEIDMLHM 111
S+ E LR++ + ++ LQ+ +E S K+ V V+++ + +L+ E+D
Sbjct: 383 SDCERLRSLCNK---KDQALQA--IESTSKKDLVETNNQVLQKLKYELKYCKGELDSAEE 547
Query: 112 TIKSLQEENNKLQEELIHEASAKRELEVARNKIKELQRQIKIIANQTKGQLLLLKQKVSG 171
TIK+L+ E L+++L KR E + + +++L+++ K + K ++ L++K+ G
Sbjct: 548 TIKTLRSEKAILEQKL--SVLEKRNSEESSSLLRKLEQERKAV----KSEVYDLERKIEG 709
Query: 172 ----LQAKEEEVVKKDAE---IENNLKTVNDF-EIEKKLKTVNDLEIEAVGLKR 217
L A + + KD+E ++NN K + + E+++ + N+ + ++R
Sbjct: 710 YRQELMAAKSIISVKDSELSALQNNFKELEELREMKEDIDRKNEQTASILKMQR 871
Score = 32.3 bits (72), Expect = 0.85
Identities = 28/115 (24%), Positives = 53/115 (45%), Gaps = 1/115 (0%)
Frame = +2
Query: 464 VSSSDDILDSVSASFQLMSKSVDRSLDEKYPAYKDLHKLALAREKQLKEKAEKARGEKFG 523
++S L+S Q +SK V+ S +KDLH+ +E +++E+ E +
Sbjct: 179 INSKPPNLESYEKRIQDLSKLVEESQRSADQLHKDLHE-KQEKEVKMQEQLEGLKESLKA 355
Query: 524 DNSNLNTTKAEEERSTSLDSELTQVNEKACVSDGLNKQS-EDVKDFDNQTITEMK 577
+ NL ++ ER SL N+K + S +D+ + +NQ + ++K
Sbjct: 356 NKQNLQAVTSDCERLRSL------CNKKDQALQAIESTSKKDLVETNNQVLQKLK 502
>TC86766 weakly similar to PIR|T06291|T06291 extensin homolog T9E8.80 -
Arabidopsis thaliana, partial (18%)
Length = 1260
Score = 38.9 bits (89), Expect = 0.009
Identities = 21/51 (41%), Positives = 24/51 (46%), Gaps = 6/51 (11%)
Frame = +2
Query: 581 IEKRPPRLPRPPPKPSDGAPVSNSLNEIPYAPSVP------SPPPPPGSLP 625
+ PP P PP P APV + +PY S P SPPPPP S P
Sbjct: 20 VRSPPPPPPNSPPPPPPPAPVFSPPPPVPYYYSSPPPPPAHSPPPPPXSPP 172
Score = 32.3 bits (72), Expect = 0.85
Identities = 20/58 (34%), Positives = 22/58 (37%)
Frame = +2
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLPRGAVGDDKVKRAPELVE 642
PP PPP P P +S P P + PPPPP P V P VE
Sbjct: 128 PPPAHSPPPPPXSPPPPPHSPTP-PVYPYLSPPPPPPVHSPPPPVYSPPPPSPPPCVE 298
Score = 31.6 bits (70), Expect = 1.4
Identities = 19/40 (47%), Positives = 21/40 (52%), Gaps = 3/40 (7%)
Frame = +2
Query: 585 PPRLPRP---PPKPSDGAPVSNSLNEIPYAPSVPSPPPPP 621
PP P P PP PS AP ++ PY P PSP PPP
Sbjct: 302 PPPPPPPCVEPPPPSSPAP-----HQTPYHPP-PSPSPPP 403
Score = 30.8 bits (68), Expect = 2.5
Identities = 16/37 (43%), Positives = 19/37 (51%), Gaps = 1/37 (2%)
Frame = +2
Query: 586 PRLPRPPPKPSDGAPVSNSLNEIPYAPSV-PSPPPPP 621
P P PPP P S + ++ PY P PSPPP P
Sbjct: 299 PPPPPPPPCVEPPPPSSPAPHQTPYHPPPSPSPPPSP 409
Score = 30.8 bits (68), Expect = 2.5
Identities = 20/46 (43%), Positives = 21/46 (45%), Gaps = 5/46 (10%)
Frame = +2
Query: 585 PPRLPRPPPKP-----SDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P S PV S P + PSPPPP S P
Sbjct: 377 PPPSPSPPPSPVYAYPSPPPPVYTSPPPSPVY-AYPSPPPPVYSSP 511
>TC85851 similar to PIR|T12180|T12180 probable transcription factor - fava
bean, partial (93%)
Length = 2007
Score = 38.9 bits (89), Expect = 0.009
Identities = 19/50 (38%), Positives = 23/50 (46%)
Frame = -3
Query: 575 EMKLFKIEKRPPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSL 624
E+ F+ + PR PRPPP P N + P P P PP PP L
Sbjct: 574 ELSSFE*SRPKPRPPRPPPNPRSPPRPPNPRSRSPPRPPNPRPPRPPSFL 425
>BQ151059 similar to GP|7291525|gb|A CG9888-PA {Drosophila melanogaster},
partial (26%)
Length = 308
Score = 38.5 bits (88), Expect = 0.012
Identities = 19/43 (44%), Positives = 19/43 (44%)
Frame = -1
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLPRG 627
PP P PPP P P P P P PPPPP PRG
Sbjct: 158 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPRG 30
Score = 38.1 bits (87), Expect = 0.015
Identities = 21/46 (45%), Positives = 21/46 (45%), Gaps = 1/46 (2%)
Frame = -3
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP-RGAV 629
PP P PPP P P P P P PPPPPG P RG V
Sbjct: 144 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPGFSPERGIV 7
Score = 38.1 bits (87), Expect = 0.015
Identities = 18/41 (43%), Positives = 18/41 (43%)
Frame = -2
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPPG P
Sbjct: 145 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPGVFP 23
Score = 35.8 bits (81), Expect = 0.077
Identities = 18/43 (41%), Positives = 18/43 (41%)
Frame = -3
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLPRG 627
PP P PPP P P P P P PPPPP P G
Sbjct: 159 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPG 31
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -1
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 293 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 171
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -3
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 288 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 166
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -1
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 302 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 180
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -3
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 279 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 157
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -1
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 296 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 174
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -3
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 276 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 154
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -2
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 274 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 152
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -2
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 271 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 149
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -3
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 267 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 145
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -2
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 265 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 143
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -2
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 256 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 134
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -3
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 282 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 160
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -1
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 209 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 87
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -3
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 252 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 130
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -1
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 251 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 129
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -1
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 248 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 126
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -2
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 244 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 122
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -3
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 240 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 118
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -2
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 238 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 116
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -1
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 236 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 114
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -1
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 233 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 111
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -1
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 230 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 108
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -1
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 227 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 105
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -1
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 224 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 102
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -3
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 219 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 97
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -2
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 217 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 95
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -1
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 260 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 138
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -1
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 212 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 90
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -3
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 213 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 91
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -3
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 204 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 82
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -1
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 203 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 81
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -1
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 200 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 78
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -3
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 195 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 73
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -2
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 193 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 71
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -1
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 191 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 69
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -1
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 188 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 66
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -1
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 185 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 63
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -2
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 181 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 59
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -1
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 179 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 57
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -1
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 176 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 54
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -2
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 172 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 50
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -3
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 168 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 46
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -2
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 166 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 44
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -2
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 163 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 41
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -1
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 263 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 141
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -1
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 287 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 165
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -3
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 297 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 175
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -2
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 304 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 182
Score = 34.3 bits (77), Expect = 0.22
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -1
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
PP P PPP P P P P P PPPPP P
Sbjct: 308 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 186
>BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (39%)
Length = 1358
Score = 38.1 bits (87), Expect = 0.015
Identities = 18/39 (46%), Positives = 21/39 (53%), Gaps = 1/39 (2%)
Frame = -1
Query: 584 RPPRLPRPPPKPSDGAPVSNSLNEIPY-APSVPSPPPPP 621
RPP P PPP P AP + + P AP +P PPP P
Sbjct: 173 RPPSAPAPPPTPPAAAPPAPTPPHAPA*APHLPPPPPAP 57
Score = 35.0 bits (79), Expect = 0.13
Identities = 19/38 (50%), Positives = 22/38 (57%)
Frame = -2
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPG 622
PP L R PP P P+++ L P A S P PPPPPG
Sbjct: 289 PPPLDRAPPPPPAPLPLASRLR--PRA-SPPPPPPPPG 185
Score = 34.7 bits (78), Expect = 0.17
Identities = 17/47 (36%), Positives = 24/47 (50%), Gaps = 5/47 (10%)
Frame = -1
Query: 580 KIEKRPPRLPRPPPKPSDGAPVSNSLNEIPYA-----PSVPSPPPPP 621
++ + P LP PP GAP + + + IP A P +P P PPP
Sbjct: 806 RVLRAPSALPAPPSATPPGAPCARAPHSIPRAHPAATPLLPPPRPPP 666
Score = 33.9 bits (76), Expect = 0.29
Identities = 21/46 (45%), Positives = 23/46 (49%), Gaps = 3/46 (6%)
Frame = -1
Query: 584 RPPRLPR---PPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLPR 626
RPPR P PPP P P S + AP P PPPP G+ PR
Sbjct: 569 RPPRPPPPVPPPPSPPRAPPQSEA------AP--PPPPPPRGARPR 456
Score = 33.5 bits (75), Expect = 0.38
Identities = 18/46 (39%), Positives = 20/46 (43%), Gaps = 6/46 (13%)
Frame = -3
Query: 585 PPRLPRPPPKPSDGAP------VSNSLNEIPYAPSVPSPPPPPGSL 624
PP PRPPP P P SL+ P P+ PPP P L
Sbjct: 171 PPLGPRPPPHPPRSGPPRPHPPTRTSLSPPPPPPAASPPPPAPPPL 34
Score = 32.0 bits (71), Expect = 1.1
Identities = 15/36 (41%), Positives = 15/36 (41%)
Frame = -2
Query: 586 PRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPP 621
PR PPP P G S P P P PPPP
Sbjct: 223 PRASPPPPPPPPGPAASAPPRPPPPPPPPPQRPPPP 116
Score = 31.6 bits (70), Expect = 1.4
Identities = 17/43 (39%), Positives = 21/43 (48%), Gaps = 1/43 (2%)
Frame = -3
Query: 584 RPPRLP-RPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
RPPR R PP+ + A + + P P PPPPP S P
Sbjct: 786 RPPRTAQRHPPRRTLRASAAQHPSRAPRRHPPPPPPPPPSSPP 658
Score = 30.8 bits (68), Expect = 2.5
Identities = 13/33 (39%), Positives = 17/33 (51%)
Frame = -2
Query: 589 PRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPP 621
PRPPP+P AP + P P+ P+ PP
Sbjct: 499 PRPPPRPPPAAPARAPPSGPPAPPTPPATRAPP 401
Score = 30.4 bits (67), Expect = 3.2
Identities = 17/48 (35%), Positives = 20/48 (41%), Gaps = 6/48 (12%)
Frame = -3
Query: 584 RPPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPS------PPPPPGSLP 625
RPP PPP P P + P P P+ PPPPP + P
Sbjct: 201 RPPPPGPPPPPPLGPRPPPHPPRSGPPRPHPPTRTSLSPPPPPPAASP 58
Score = 30.0 bits (66), Expect = 4.2
Identities = 22/64 (34%), Positives = 26/64 (40%), Gaps = 22/64 (34%)
Frame = -1
Query: 584 RPPRLPRPPPKPSDG---APVSNSLNEIPYA------------------PSVPSPPP-PP 621
RPP RPP P G AP L++ P A PS P+PPP PP
Sbjct: 314 RPPPPTRPPAAPGPGPPPAPGPTPLSKPPAAEGIPPATAPPPRARRLRPPSAPAPPPTPP 135
Query: 622 GSLP 625
+ P
Sbjct: 134 AAAP 123
Score = 30.0 bits (66), Expect = 4.2
Identities = 17/56 (30%), Positives = 21/56 (37%), Gaps = 14/56 (25%)
Frame = -1
Query: 585 PPRLPRPPPKPSDGAPVSNSLN--------------EIPYAPSVPSPPPPPGSLPR 626
PP P PPP + P++ + P P P P PPP S PR
Sbjct: 686 PPPRPPPPPPEWERVPLAKKIGGQPRPALVPLVRVATYPRPPRPPPPVPPPPSPPR 519
Score = 30.0 bits (66), Expect = 4.2
Identities = 17/45 (37%), Positives = 19/45 (41%), Gaps = 1/45 (2%)
Frame = -2
Query: 584 RPPRLPRPPP-KPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLPRG 627
RPP P PPP +P P E P +P PP P P G
Sbjct: 163 RPPPPPPPPPQRPPPPPPPHTHQPEPPTSPPRRQPPAPRAPPPSG 29
Score = 29.3 bits (64), Expect = 7.2
Identities = 16/43 (37%), Positives = 20/43 (46%)
Frame = -2
Query: 583 KRPPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
+RPP RP P P ++ E A +PPPPP S P
Sbjct: 793 RRPPSPHRPAPPPQ-----AHPARERRTASLARTPPPPPSSPP 680
Score = 28.9 bits (63), Expect = 9.4
Identities = 17/41 (41%), Positives = 17/41 (41%)
Frame = -1
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLP 625
P LPRPPP P AP P PPP PG P
Sbjct: 329 PRPLPRPPPP-----------TRPPAAPG-PGPPPAPGPTP 243
>TC77148 ENOD20
Length = 1108
Score = 38.1 bits (87), Expect = 0.015
Identities = 32/102 (31%), Positives = 42/102 (40%), Gaps = 11/102 (10%)
Frame = +3
Query: 576 MKLFKIEKRPPRLPRPPPKPSDGAPVSNSLNEIPYAP--SVPSPP---------PPPGSL 624
+KL + P L PPP PS P S IP+ P S+PSPP P P
Sbjct: 381 LKLAVVVMVAPVLSSPPPPPS--PPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPS 554
Query: 625 PRGAVGDDKVKRAPELVEFYQSLMKREAKKDASLLTSSTSNA 666
PR KR+P SL K + ++ L S S++
Sbjct: 555 PRSTPIPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDS 680
>TC84030
Length = 664
Score = 38.1 bits (87), Expect = 0.015
Identities = 35/123 (28%), Positives = 48/123 (38%), Gaps = 3/123 (2%)
Frame = +2
Query: 586 PRLPRPPPKPSDGAPVSNS--LNEIP-YAPSVPSPPPPPGSLPRGAVGDDKVKRAPELVE 642
P P P P PSDGA + S IP APS+ SP P P G D +
Sbjct: 95 PYYPYPTPAPSDGASFARSSYAGYIPSEAPSLASPLPKSTDFP--GYGSDYL-------- 244
Query: 643 FYQSLMKREAKKDASLLTSSTSNAADTRSNVIAEIENRSSFLLAVKADVETQGDFVMSLA 702
KD SL DTR + + N +S+ D+ T+ D ++ ++
Sbjct: 245 ----------NKDVSLFRMEPYGVDDTRGSRVHSEHNATSYNPLEDVDLSTKRDALLGVS 394
Query: 703 TEV 705
T V
Sbjct: 395 TGV 403
>BQ140560 weakly similar to GP|15128223|db hypothetical protein~similar to
Oryza sativa chromosome 1 P0489A01.2, partial (5%)
Length = 215
Score = 38.1 bits (87), Expect = 0.015
Identities = 19/42 (45%), Positives = 23/42 (54%), Gaps = 3/42 (7%)
Frame = +1
Query: 587 RLPRPPPKPSDGAPVSNSLNEIPY---APSVPSPPPPPGSLP 625
+ P PPP P GAP + +P AP P PPPPP S+P
Sbjct: 10 KAPPPPPPPFYGAPPPPPPSYLPASSGAPLPPPPPPPPPSMP 135
Score = 30.0 bits (66), Expect = 4.2
Identities = 16/36 (44%), Positives = 18/36 (49%)
Frame = +1
Query: 585 PPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPP 620
PP +P PPP P GAP L Y P+ PPP
Sbjct: 121 PPSMP-PPPPPFHGAPPPPXL----YLPASTGAPPP 213
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.310 0.128 0.345
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,110,811
Number of Sequences: 36976
Number of extensions: 306788
Number of successful extensions: 5567
Number of sequences better than 10.0: 277
Number of HSP's better than 10.0 without gapping: 2897
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4294
length of query: 915
length of database: 9,014,727
effective HSP length: 105
effective length of query: 810
effective length of database: 5,132,247
effective search space: 4157120070
effective search space used: 4157120070
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146570.7


